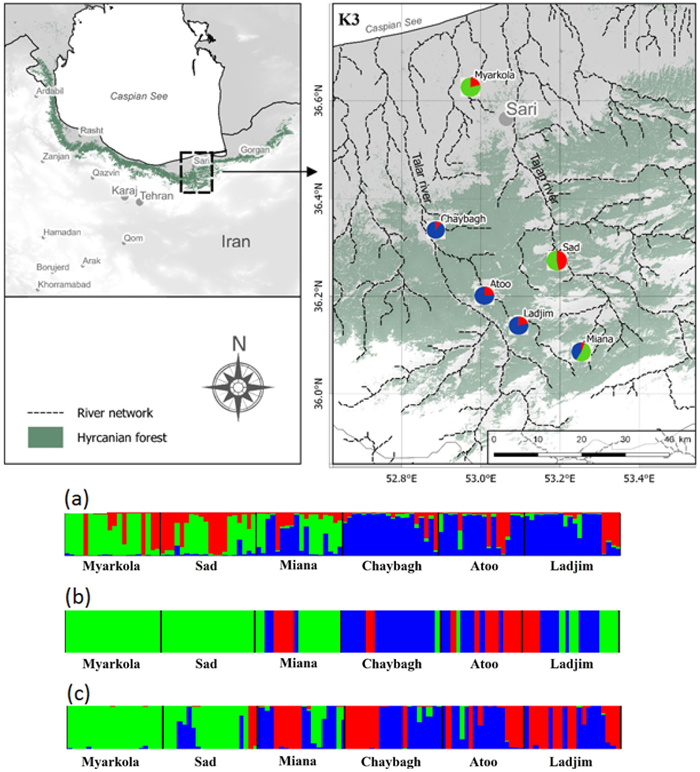
Fig. 1. Bayesian clustering results for Pterocarya fraxinifolia in the Hyrcanian region (N = 116) estimated along two altitudinal gradients from K = 3 and an associated map of the mean membership probabilities per site from STRUCTURE. Each individual is represented by a vertical bar partitioned into K segments, which represents the amount of ancestry of its genome corresponding to K clusters. Visualization was improved by sorting the genotypes by site. Gradient 1: Myarkola, Sad, and Miana; gradient 2: Chaybagh, Atoo, and Ladjim. (a) Output from STRUCTURE. (b) Output from BAPS. (c) Results of DAPC analysis in R.
| Table 1. Geographical characteristics and sample size of investigated populations in two gradients along two river systems. | |||||
| Population name | Latitude (UTM) | Longitude (UTM) | Altitude (m a.s.l.) | Sample size | |
| Tajan river (gradient 1) | Myarkola | 672536.3 | 4051845.08 | 50–60 | 20 |
| Sad | 697694.9 | 4017597.8 | 350–400 | 20 | |
| Miana | 703567.39 | 3996839.29 | 900–970 | 18 | |
| Talar river (gradient 2) | Chaybagh | 669888.3 | 4023492.3 | 200–250 | 20 |
| Atoo | 681346.47 | 4008915.14 | 400–450 | 18 | |
| Ladjim | 686691.74 | 4013147.26 | 900–950 | 20 | |
| Table 2. Genetic variability within two gradients of Pterocarya fraxinifolia populations. | |||||||
| Elevation | Population | Effective number of alleles (Ne) | Observed heterozygosity (Ho) | Expected heterozygosity (He) | Allelic richness (Ar) | Private allelic richness (PAr) | |
| Gradient 1 | Low | Myarkola | 5.7 ± 0.73 | 0.78 ± 0.1 | 0.807 ± 0.028 | 4.06 | 0.99 |
| Medium | Sad | 4.7 ± 0.36 | 0.85 ± 0.067 | 0.78 ± 0.02 | 3.8 | 0.4 | |
| High | Miana | 5.1 ± 0.44 | 0.86 ± 0.07 | 0.79 ± 0.02 | 3.9 | 0.38 | |
| Gradient 2 | Low | Chaybagh | 3.72 ± 0.23 | 0.79 ± 0.11 | 0.726 ± 0.018 | 3.3 | 0.1 |
| Medium | Atoo | 4.27 ± 0.51 | 0.87 ± 0.09 | 0.751 ± 0.025 | 3.5 | 0.2 | |
| High | Ladjim | 4.25 ± 0.69 | 0.86 ± 0.11 | 0.730 ± 0.043 | 3.4 | 0.14 | |
| Table 3. Analyses of molecular variance (AMOVA) for two gradients of Pterocarya fraxinifolia from Hyrcanian forest using SSR markers. | ||||||||||
| AMOVA component | AMOVA genetic distance option | |||||||||
| S.O.V | d.f | SS | MS | Est. Var. | % of Variance | PhiPT | Rst | Fst | Nm | |
| Gradient 1 | Among Pops | 2 | 27.76 | 13.8 | 0.45 | 9% | 0.086** | 0.049** | 0.046** | 5.13 |
| Within Pops | 55 | 284.95 | 5.18 | 5.18 | 91% | |||||
| Total | 57 | 312.72 | 5.63 | 100% | ||||||
| Gradient 2 | Among Pops | 2 | 22.13 | 11.06 | 0.34 | 7 | 0.075** | 0.052** | 0.036** | 6.6 |
| Within Pops | 55 | 237.68 | 4.32 | 4.32 | 93 | |||||
| Total | 57 | 259.82 | 4.67 | 100 | ||||||
| Total | Among Pops | 5 | 85.1 | 17 | 0.63 | 12 | 0.11** | 0.037** | 0.062** | 3.77 |
| Within | 110 | 521.3 | 4.7 | 88 | ||||||
| Total | 115 | 606.4 | 4.7 | 5.37 | 100 | |||||
| Statistics include sums of squared deviations (SS); mean squared deviations (MS), variance component estimates (Est. Var.). ** – p-value < 0.01. | ||||||||||
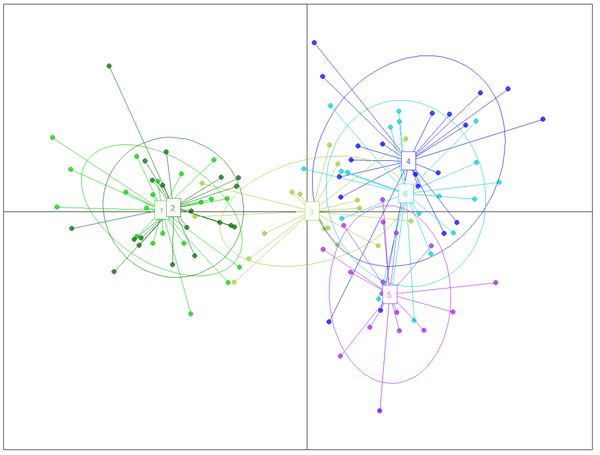
Fig. 2. The result of DAPC analysis with populations used as clusters: 1 – Myarkola, 2 – Sad, 3 – Miana, 4 – Chaybagh, 5 – Atoo, 6 – Ladjim. Green colours indicated populations from gradient 1, blue and purple – populations from gradient 2.
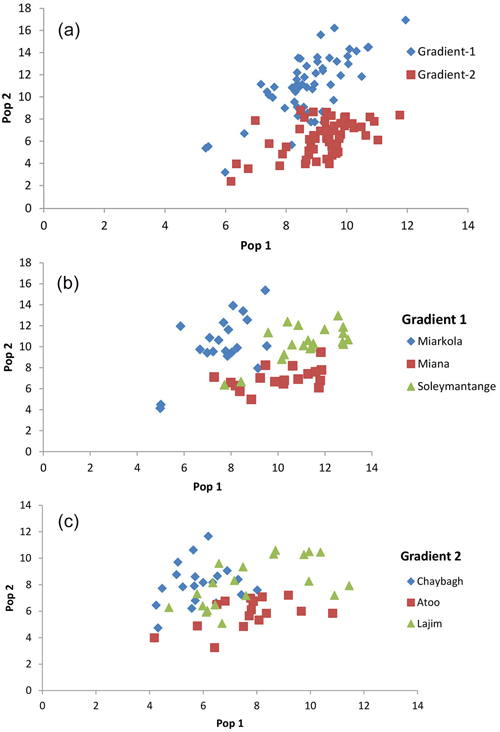
Fig. 3. Result of the population assignment. (a) Pairwise population assignment graphs of gradient 1 vs. gradient 2. (b) Assignment of the populations of gradient 1. (c) Assignment of the populations of gradient 2.
| Table 4. Summary of population assignment outcomes to “self” or “other” population. | ||||||
| Pop | Gradient 1 Pops | Gradient 2 Pops | Total | Percent | ||
| All populations* | Self Pop | 52 | 57 | 109 | 94% | |
| Other Pop | 6 | 1 | 7 | 6% | ||
| Pop | Myarkola | Sad | Miana | Total | Percent | |
| Gradient 1 | Self Pop | 17 | 20 | 18 | 55 | 95% |
| Other Pop | 3 | 0 | 0 | 3 | 5% | |
| Pop | Chaybagh | Atoo | Ladjim | Total | Percent | |
| Gradient 2 | Self Pop | 18 | 16 | 16 | 50 | 86% |
| Other Pop | 2 | 2 | 4 | 8 | 14% | |
| * – All population in each gradient grouped as one population. | ||||||
| Table 5. Pairwise population gene flow (Nm) values (below diagonal) and pairwise population genetic distance based on Fst values (above diagonal). | ||||
| Populations | Myarkola | Sad | Miana | |
| Gradient 1 | Myarkola | 0.000 | 0.058 | 0.025 |
| Sad | 9.88 | 0.000 | 4.343 | |
| Miana | 4.343 | 0.054 | 0.000 | |
| Populations | Chaybagh | Atoo | Ladjim | |
| Gradient 2 | Chaybagh | 0.000 | 0.041 | 0.026 |
| Atoo | 5.856 | 0.000 | 0.043 | |
| Ladjim | 9.310 | 5.564 | 0.000 | |
| Nm – the product of the effective population number and rate of migration among populations. | ||||
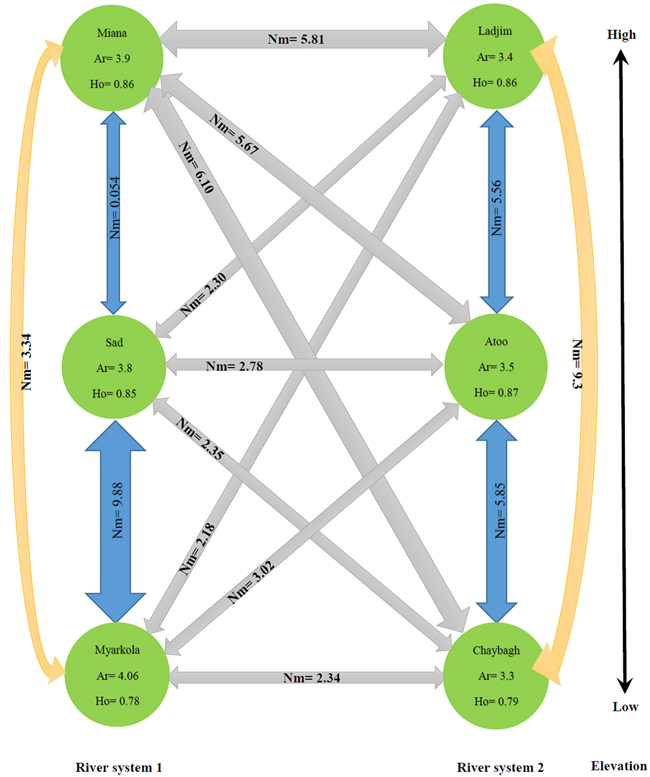
Fig. 4. Migration pattern of Pterocarya fraxinifolia within and among the river populations.