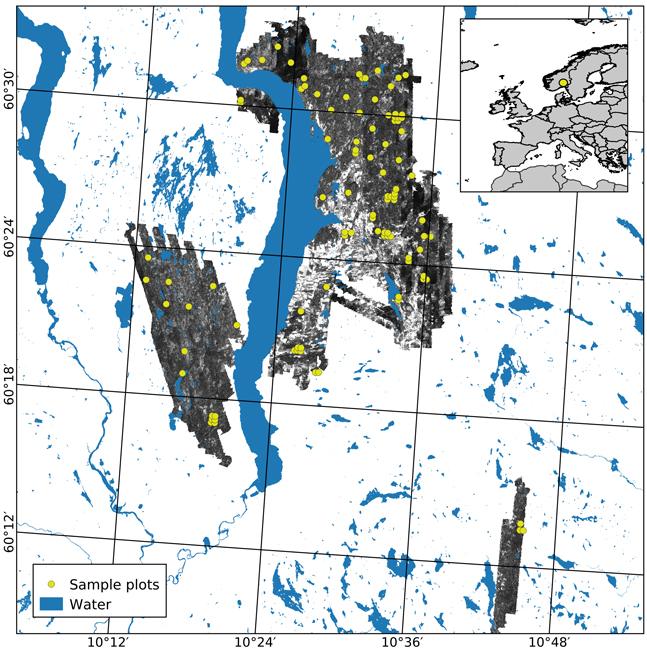
Fig. 1. The geographical location of the study area in Norway (insert) and the extent of the hyperspectral acquisition are shown using an image from the NIR band (708 nm). Sample plots appear in yellow.
| Table 1. Summary of volume and species proportions on the 97 sample plots used for training and validation for Spruce (Picea abies), Pine (Pinus sylvestris), Deciduous species (mainly birch (Betula spp.)) and in total. Mean values for all plots are presented in parenthesis. | ||
| Species | Species proportion, (mean) (%) | Volume, (mean) in m3 ha–1 |
| Spruce | 1–100 (67) | 1–625 (168) |
| Pine | 0–98 (21) | 0–369 (48) |
| Deciduous | 0–83 (12) | 0–155 (25) |
| Total | 16–625 (241) | |
| Table 2. Summary of number of plots, percent of dominant species of total plot volume and volume for training and validation plots by dominant species; Spruce-dominated (Picea abies), Pine-dominated (Pinus sylvestris), Deciduous-dominated (mainly dominated by Birch (Betula spp.)) and in total. Mean values for all plots are presented in parenthesis. | |||
| Dominant species | Number of plots | % of dominant species of total plot volume (mean) | Volume, (mean) in m3 ha–1 |
| Spruce-dominated | 70 | 45–100 (85) | 24–625 (257) |
| Pine-dominated | 22 | 39–98 (71) | 65–471 (227) |
| Deciduous-dominated | 5 | 53–83 (68) | 16–266 (129) |
| Total | 97 | 16–625 (241) | |
| Table 3. Computation and reference for vegetation indices derived from hyperspectral data and applied in the modelling of species composition. | ||
| Index | Computation | Reference |
| Normalized Difference Vegetation Index | Rouse et al. (1973) | |
| Renormalized Difference Vegetation Index | Roujean and Breon (1995) | |
| Modified red edge Simple Ratio index | Chen (1996) | |
| Conifer Index | Trier et al. (2018) | |
| Spruce Index | Trier et al. (2018) | |
| Soil-Adjusted Vegetation Index | Huete (1988) | |
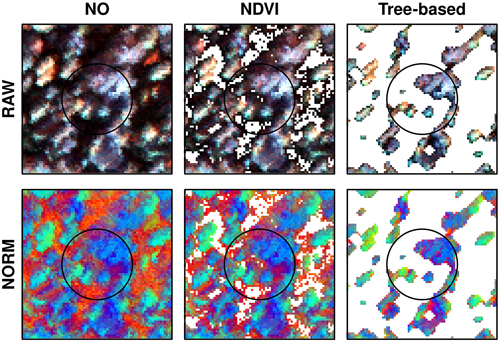
Fig. 2. Example from one field plot applying different normalization (RAW, NORM) and thresholding approaches (NO, NDVI, tree-based). Pixels removed applying thresholding appear white and are not used when computing variables for modelling. Image colors: red = 445 nm, green=714 nm, and blue=839 nm using a linear stretch.
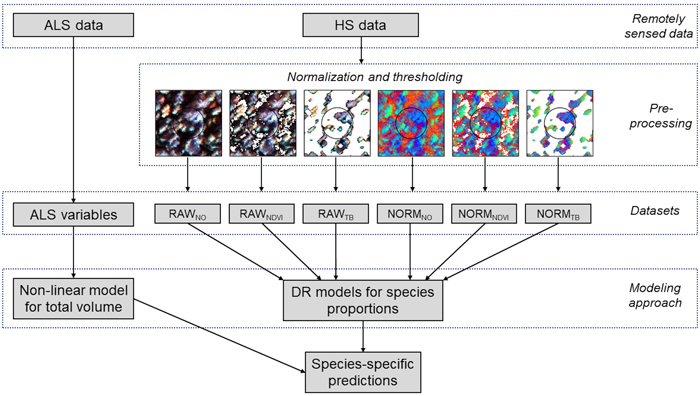
Fig. 3. Schematic diagram of the data processing and modeling steps used in the study. From the remotely sensed data preprocessing is applied to the hyperspectral (HS) data in terms of normalization and thresholding, providing six different datasets for Dirichlet (DR) modelling of species proportions; RAWNO (no normalization and no thresholding), RAWNDVI (no normalization and NDVI thresholding), RAWTB (no normalization and tree-based (TB) thresholding), NORMNO (normalization and no thresholding), NORMNDVI (normalization and NDVI thresholding), and NORMTB (normalization and TB thresholding). Variables from airborne laser scanning (ALS) data are used in modeling of total volume.
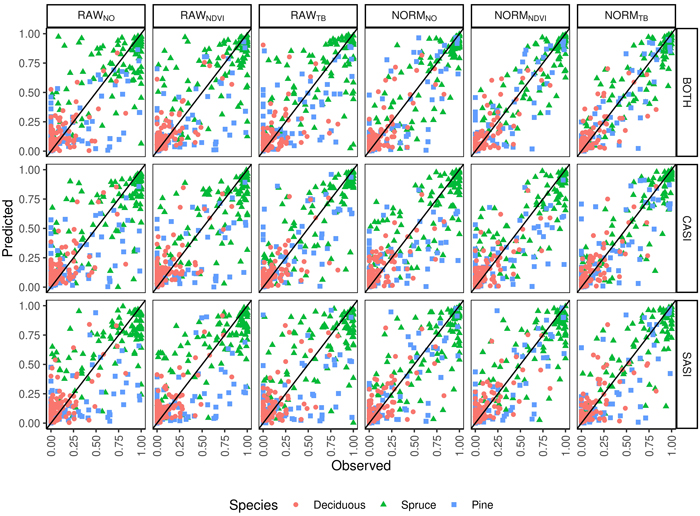
Fig. 4. Observed and predicted species proportions (Spruce = Norway spruce (Picea abies), Pine = Scots pine (Pinus sylvestris), Deciduous (mainly birch (Betula spp.))) for different sensors (CASI, SASI) and their combination (BOTH) for different preprocessing steps (RAWNO, RAWNDVI, RAWTB, NORMNO, NORMNDVI, NORMTB).
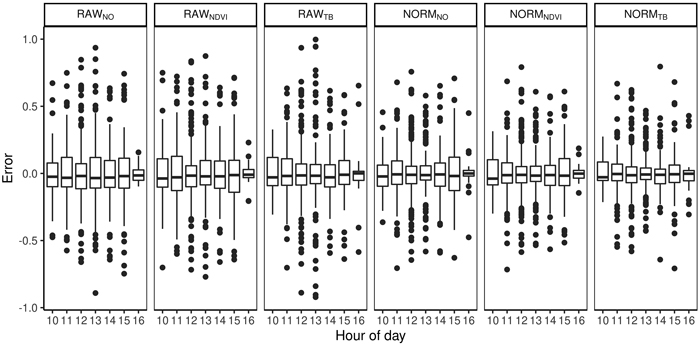
Fig. 5. Difference between predicted and observed species proportions for different hours of the day and for different preprocessing steps (RAWNO, RAWNDVI, RAWTB, NORMNO, NORMNDVI, NORMTB).
| Table 4. Results for species-specific volume and fuzzy set accuracy for the Dirichlet regression approach. OA = overall accuracy, RRMSDS = relative root mean squared error for Norway spruce (Picea abies), RRMSDP = relative root mean squared error for Scots pine (Pinus sylvestris), RRMSDD = relative root mean squared error for deciduous trees (mainly birch (Betula spp.)). | |||||||||||
| Data | Sensor | Kappa-value | OA | RRMSDS | RRMSDP | RRMSDD | Fuzzy set accuracy | ||||
| Five | Four | Three | Two | One | |||||||
| RAWNO | CASI | 0.53 | 0.81 | 48 | 142.3 | 142.6 | 50 | 73 | 85 | 15 | 11 |
| RAWNO | SASI | 0.39 | 0.77 | 42.6 | 150.6 | 139.7 | 45 | 74 | 83 | 18 | 11 |
| RAWNO | BOTH | 0.53 | 0.81 | 46.6 | 139.5 | 128 | 53 | 76 | 85 | 16 | 10 |
| RAWNDVI | CASI | 0.69 | 0.87 | 47.2 | 148.3 | 140.8 | 51 | 75 | 86 | 13 | 8 |
| RAWNDVI | SASI | 0.43 | 0.79 | 41.2 | 148.6 | 134.8 | 47 | 72 | 86 | 14 | 9 |
| RAWNDVI | BOTH | 0.55 | 0.82 | 46.6 | 146.6 | 114.7 | 57 | 76 | 86 | 14 | 10 |
| RAWTB | CASI | 0.61 | 0.84 | 41.2 | 120.2 | 107.5 | 61 | 83 | 90 | 10 | 6 |
| RAWTB | SASI | 0.57 | 0.82 | 48.6 | 146.3 | 154.4 | 48 | 72 | 85 | 15 | 10 |
| RAWTB | BOTH | 0.52 | 0.8 | 48.1 | 134.7 | 123.5 | 57 | 78 | 86 | 14 | 10 |
| NORMNO | CASI | 0.42 | 0.77 | 37.8 | 135.8 | 124.6 | 53 | 73 | 85 | 16 | 8 |
| NORMNO | SASI | 0.66 | 0.86 | 40.6 | 114.2 | 148.9 | 58 | 78 | 87 | 12 | 6 |
| NORMNO | BOTH | 0.66 | 0.85 | 35 | 117.6 | 117.6 | 64 | 81 | 90 | 9 | 5 |
| NORMNDVI | CASI | 0.62 | 0.85 | 35.4 | 131.5 | 129.4 | 60 | 76 | 87 | 13 | 5 |
| NORMNDVI | SASI | 0.65 | 0.86 | 37.7 | 120.6 | 124 | 58 | 81 | 91 | 8 | 5 |
| NORMNDVI | BOTH | 0.79 | 0.91 | 32.4 | 104.9 | 109.6 | 68 | 84 | 93 | 6 | 2 |
| NORMTB | CASI | 0.76 | 0.9 | 36.6 | 105 | 100 | 66 | 85 | 92 | 8 | 4 |
| NORMTB | SASI | 0.69 | 0.87 | 38.1 | 122.9 | 132.3 | 64 | 82 | 92 | 8 | 6 |
| NORMTB | BOTH | 0.70 | 0.88 | 34.2 | 87 | 102.2 | 68 | 88 | 93 | 6 | 2 |
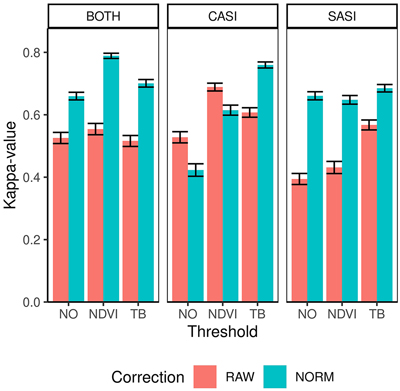
Fig. 6. Kappa values and confidence intervals for dominant species obtained by different sensors (CASI and SASI) and their combination (BOTH). In addition, the different preprocessing steps, i.e., using raw data (RAW) or normalized data (NORM) and different thresholding methods (NO, NDVI or TB).
| Table 5. Error matrices of the dominant species classification (S = Norway spruce (Picea abies), P = Scots pine (Pinus sylvestris), D = deciduous (mainly birch (Betula spp.))) obtained from the Dirichlet regression for different sensors (CASI, SASI and combined BOTH) and preprocessing methods (Data). PA = producer’s accuracy and UA = User’s accuracy. | |||||||||||||
| Data | Sensor | CASI | SASI | BOTH | |||||||||
| Species | S | P | D | PA | S | P | D | PA | S | P | D | PA | |
| RAWNO | S | 65 | 11 | 1 | 84.4 | 66 | 13 | 2 | 81.5 | 65 | 10 | 2 | 84.4 |
| P | 3 | 10 | 0 | 76.9 | 2 | 6 | 0 | 75.0 | 3 | 11 | 0 | 78.6 | |
| D | 2 | 1 | 4 | 57.1 | 2 | 3 | 3 | 37.5 | 2 | 1 | 3 | 50.0 | |
| UA | 92.9 | 45.5 | 80.0 | 81.4 | 94.3 | 27.3 | 60.0 | 77.3 | 92.9 | 50.0 | 60.0 | 81.4 | |
| RAWNDVI | S | 64 | 6 | 0 | 91.4 | 67 | 15 | 1 | 80.7 | 65 | 12 | 0 | 84.4 |
| P | 4 | 15 | 0 | 78.9 | 1 | 6 | 0 | 85.7 | 3 | 10 | 0 | 76.9 | |
| D | 2 | 1 | 5 | 62.5 | 2 | 1 | 4 | 57.1 | 2 | 0 | 5 | 71.4 | |
| UA | 91.4 | 68.2 | 100.0 | 86.6 | 95.7 | 27.3 | 80.0 | 79.4 | 92.9 | 45.5 | 100.0 | 82.5 | |
| RAWTB | S | 63 | 6 | 2 | 88.7 | 65 | 9 | 1 | 86.7 | 63 | 8 | 3 | 85.1 |
| P | 6 | 15 | 0 | 71.4 | 3 | 11 | 0 | 78.6 | 5 | 13 | 0 | 72.2 | |
| D | 1 | 1 | 3 | 60.0 | 2 | 2 | 4 | 50.0 | 2 | 1 | 2 | 40.0 | |
| UA | 90.0 | 68.2 | 60.0 | 83.5 | 92.9 | 50.0 | 80.0 | 82.5 | 90.0 | 59.1 | 40.0 | 80.4 | |
| NORMNO | S | 63 | 13 | 1 | 81.8 | 64 | 5 | 2 | 90.1 | 61 | 4 | 1 | 92.4 |
| P | 5 | 8 | 0 | 61.5 | 2 | 16 | 0 | 88.9 | 4 | 17 | 0 | 81.0 | |
| D | 2 | 1 | 4 | 57.1 | 4 | 1 | 3 | 37.5 | 5 | 1 | 4 | 40.0 | |
| UA | 90.0 | 36.4 | 80.0 | 77.3 | 91.4 | 72.7 | 60.0 | 85.6 | 87.1 | 77.3 | 80.0 | 84.5 | |
| NORMNDVI | S | 65 | 9 | 1 | 86.7 | 65 | 5 | 3 | 89.0 | 66 | 3 | 0 | 95.7 |
| P | 3 | 13 | 0 | 81.2 | 3 | 16 | 0 | 84.2 | 1 | 17 | 0 | 94.4 | |
| D | 2 | 0 | 4 | 66.7 | 2 | 1 | 2 | 40.0 | 3 | 2 | 5 | 50.0 | |
| UA | 92.9 | 59.1 | 80.0 | 84.5 | 92.9 | 72.7 | 40.0 | 85.6 | 94.3 | 77.3 | 100.0 | 90.7 | |
| NORMTB | S | 66 | 3 | 1 | 94.3 | 65 | 4 | 2 | 91.5 | 66 | 5 | 2 | 90.4 |
| P | 3 | 17 | 0 | 85.0 | 2 | 16 | 0 | 88.9 | 1 | 16 | 0 | 94.1 | |
| D | 1 | 2 | 4 | 57.1 | 3 | 2 | 3 | 37.5 | 3 | 1 | 3 | 42.9 | |
| UA | 94.3 | 77.3 | 80.0 | 89.7 | 92.9 | 72.7 | 60.0 | 86.6 | 94.3 | 72.7 | 60.0 | 87.6 | |
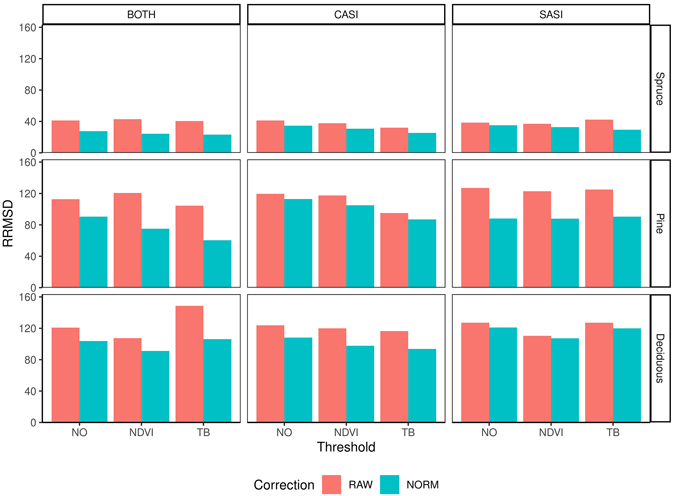
Fig. 7. Relative root mean square difference (RRMSD) for different preprocessing steps, i.e., using raw data (RAW) or normalized data (NORM), and thresholding methods (NO, NDVI, TB), species (Spruce = Norway spruce (Picea abies), Pine = Scots pine (Pinus sylvestris), Deciduous (mainly birch (Betula spp.))), sensors (CASI and SASI) and their combination (BOTH).
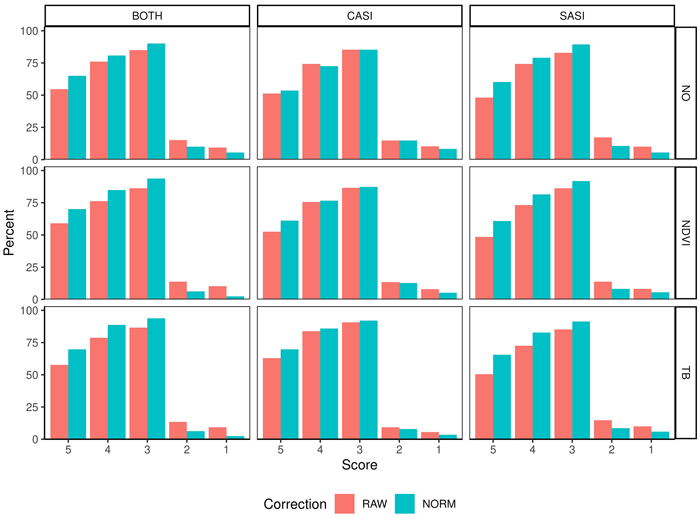
Fig. 8. Percentage of observations in different score values (5–1) obtained from fuzzy validation by different sensors (CASI and SASI) and their combination (BOTH). In addition, the different preprocessing steps, i.e., using raw data (RAW) or normalized data (NORM) and different thresholding methods (NO, NDVI or TB). A score of 5 indicates no deviation between predicted and observed species proportion and a lower score indicates subsequent higher deviation; for details see section 2.4.3.
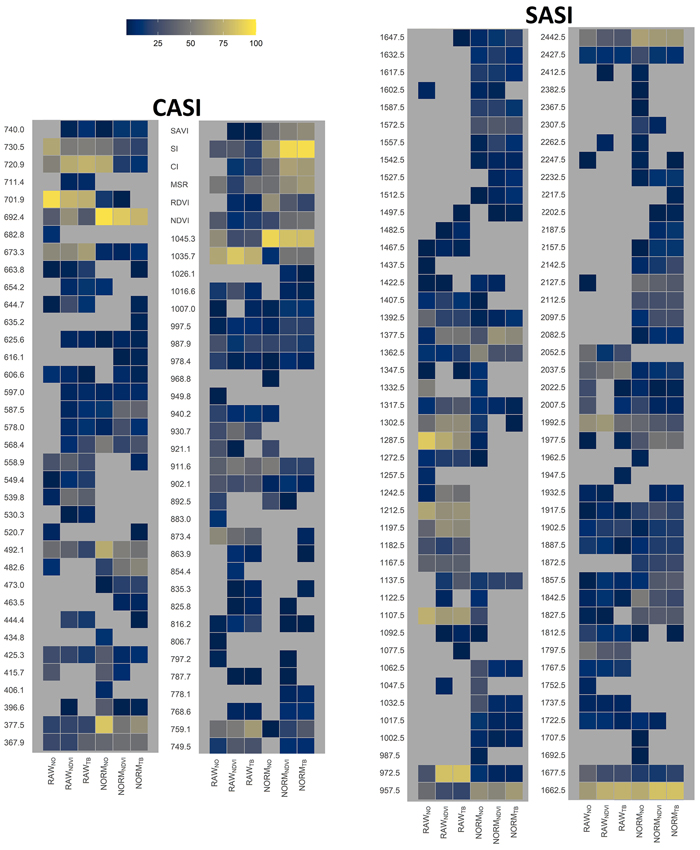
Fig. 9. Relative importance of HS-derived variables, i.e., vegetation index name or wavelength obtained by different sensors (CASI and SASI) and the different preprocessing steps, i.e., using raw data (RAW) or normalized data (NORM) and different thresholding methods (NO, NDVI or TB). Importance was computed as the percentage the variable was selected in the cross-validation. View larger in new window/tab.