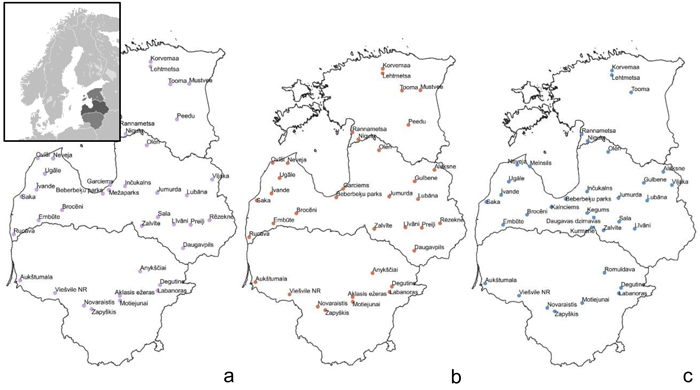
Fig. 1. Sample collection sites within the Baltic states – Estonia, Latvia, Lithuania (from top to bottom) to assess genetic diversity and structure of Vaccinium myrtillus (a), V. vitis-idaea (b) and V. uliginosum (c). View larger in new window/tab.
| Table 1. EST-SSR markers used for Vaccinium species analysis. X – marker used in the respective species. | |||||
| Locus | Annealing temperature, °C | Allele size range (bp) | V. myrtillus | V. vitis-idaea | V. uliginosum |
| NA961 | 60 | 174–189 | x | x | |
| CA794F | 58–60 | 238–270 | x | x | |
| CA483F | 58 | 312–322 | x | ||
| CA112F | 58–60 | 170–200 | x | x | x |
| NA741 | 58–60 | 316–360 | x | x | x |
| CA236F | 60–62 | 227–247 | x | x | |
| CA421F | 58–60 | 164–212 | x | x | x |
| VCC_K4 | 60 | 199–265 | x | x | |
| VCC_J9 | 60 | 106–188 | x | x | |
| NA398 | 58 | 219–241 | x | ||
| NA1040 | 60 | 205–247 | x | ||
| CA344F | 60 | 124–184 | x | ||
| CA642F | 60 | 302–354 | x | ||
| VCC_I2 | 60 | 205–219 | x | x | |
| Table 2. Genetic diversity parameters, obtained using EST-SSR markers in the analysed Vaccinium myrtillus populations. | |||||||||
| Population number | Population | Coordinates (N, E) | No. of indivi- duals | Average no. of alleles (SE) | Average no. of effective alleles (SE) | Average Information index (SE) | Average Unbiased Expected heterozy- gosity (SE) | Average Observed heterozy- gosity (SE) | No. of unique frag- ments |
| Latvia | |||||||||
| 1 | Zalvīte | 56.372, 25.208 | 24 | 4.00 (1.20) | 2.31 (0.43) | 0.89 (0.20) | 0.48 (0.08) | 0.36 (0.10) | 0 |
| 2 | Ugāle | 57.289, 22.047 | 24 | 4.50 (1.27) | 2.09 (0.29) | 0.83 (0.19) | 0.45 (0.09) | 0.39 (0.10) | 1 |
| 3 | Neveja | 57.564, 22.307 | 24 | 3.75 (1.08) | 2.04 (0.31) | 0.78 (0.20) | 0.42 (0.09) | 0.39 (0.10) | 0 |
| 4 | Rucava | 56.195, 21.107 | 24 | 3.88 (0.95) | 1.98 (0.27) | 0.76 (0.16) | 0.42 (0.09) | 0.32 (0.09) | 1 |
| 5 | Oleri | 57.802, 25.414 | 24 | 3.50 (0.87) | 2.10 (0.33) | 0.80 (0.18) | 0.45 (0.08) | 0.39 (0.10) | 0 |
| 6 | Lubāna | 56.892, 26.682 | 24 | 4.00 (1.18) | 2.06 (0.33) | 0.78 (0.18) | 0.43 (0.09) | 0.32 (0.09) | 0 |
| 7 | Jumurda | 56.955, 25.725 | 24 | 4.38 (1.25) | 2.13 (0.30) | 0.86 (0.19) | 0.46 (0.08) | 0.40 (0.09) | 0 |
| 8 | Sala | 56.524, 25.727 | 18 | 4.75 (1.26) | 2.61 (0.68) | 0.98 (0.22) | 0.50 (0.09) | 0.45 (0.11) | 2 |
| 9 | Garciems | 57.110, 24.193 | 24 | 3.50 (0.76) | 1.94 (0.26) | 0.76 (0.17) | 0.42 (0.09) | 0.30 (0.07) | 0 |
| 10 | Inčukalns | 57.084, 24.676 | 24 | 4.00 (1.20) | 2.38 (0.46) | 0.86 (0.23) | 0.47 (0.11) | 0.46 (0.11) | 0 |
| 11 | Preiļi | 56.368, 26.771 | 24 | 3.50 (0.57) | 1.93 (0.25) | 0.75 (0.14) | 0.42 (0.08) | 0.41 (0.10) | 0 |
| 12 | Līvāni | 56.386, 26.235 | 23 | 3.50 (0.80) | 1.95 (0.21) | 0.77 (0.13) | 0.45 (0.06) | 0.44 (0.08) | 0 |
| 13 | Rēzekne | 56.434, 27.378 | 24 | 3.13 (0.64) | 1.86 (0.21) | 0.69 (0.15) | 0.40 (0.09) | 0.35 (0.11) | 0 |
| 14 | Daugavpils | 55.956, 26.497 | 26 | 4.50 (1.27) | 2.20 (0.46) | 0.84 (0.22) | 0.43 (0.10) | 0.40 (0.10) | 1 |
| 15 | Embūte | 56.461, 21.887 | 24 | 3.75 (0.82) | 2.26 (0.26) | 0.91 (0.15) | 0.51 (0.07) | 0.42 (0.06) | 0 |
| 16 | Brocēni | 56.654, 22.638 | 24 | 4.38 (1.02) | 2.23 (0.35) | 0.89 (0.18) | 0.48 (0.08) | 0.40 (0.09) | 0 |
| 17 | Saka | 56.869, 21.304 | 24 | 4.63 (1.27) | 2.26 (0.34) | 0.93 (0.19) | 0.50 (0.08) | 0.49 (0.08) | 0 |
| 18 | Ovīši | 57.564, 21.797 | 24 | 4.00 (1.20) | 2.26 (0.41) | 0.83 (0.23) | 0.44 (0.11) | 0.44 (0.12) | 0 |
| 19 | Īvande | 57.004, 21.792 | 24 | 3.75 (0.65) | 2.16 (0.29) | 0.85 (0.15) | 0.47 (0.08) | 0.32 (0.10) | 0 |
| 20 | Viļaka | 57.155, 27.537 | 23 | 3.63 (0.53) | 2.08 (0.26) | 0.81 (0.15) | 0.46 (0.08) | 0.32 (0.10) | 0 |
| 21 | Beberbeķu parks | 56.952, 23.957 | 24 | 4.25 (0.75) | 2.28 (0.38) | 0.89 (0.18) | 0.48 (0.09) | 0.45 (0.10) | 0 |
| 22 | Mežaparks | 57.012, 24.157 | 24 | 4.25 (1.03) | 2.40 (0.40) | 0.91 (0.20) | 0.50 (0.09) | 0.40 (0.10) | 0 |
| Estonia | |||||||||
| 23 | Rannametsa | 58.134, 24.512 | 16 | 3.13 (0.44) | 2.37 (0.31) | 0.87 (0.15) | 0.52 (0.09) | 0.41 (0.08) | 0 |
| 24 | Lehtmetsa | 59.203, 25.600 | 16 | 3.63 (0.98) | 2.24 (0.37) | 0.85 (0.20) | 0.47 (0.09) | 0.37 (0.10) | 0 |
| 25 | Mustvee | 58.870, 26.919 | 16 | 3.50 (0.73) | 2.02 (0.30) | 0.77 (0.18) | 0.43 (0.10) | 0.32 (0.09) | 0 |
| 26 | Nigula | 58.007, 24.714 | 16 | 3.50 (0.76) | 1.91 (0.26) | 0.74 (0.15) | 0.42 (0.08) | 0.36 (0.09) | 0 |
| 27 | Korvemaa | 59.285, 25.611 | 16 | 4.25 (0.90) | 2.21 (0.40) | 0.85 (0.21) | 0.44 (0.10) | 0.34 (0.10) | 0 |
| 28 | Tooma | 58.876, 26.272 | 16 | 3.38 (0.84) | 1.91 (0.27) | 0.74 (0.17) | 0.41 (0.08) | 0.37 (0.10) | 0 |
| 29 | Peedu | 58.248, 26.453 | 17 | 3.38 (0.84) | 2.14 (0.36) | 0.77 (0.19) | 0.44 (0.10) | 0.35 (0.10) | 0 |
| Lithuania | |||||||||
| 30 | Motiejunai village | 55.051, 24.485 | 13 | 2.88 (0.55) | 1.97 (0.27) | 0.72 (0.17) | 0.43 (0.09) | 0.43 (0.12) | 0 |
| 31 | Anykščiai | 55.566, 25.126 | 16 | 3.88 (0.92) | 2.20 (0.26) | 0.87 (0.16) | 0.49 (0.09) | 0.36 (0.09) | 0 |
| 32 | Degutine forest | 55.315, 25.738 | 16 | 3.75 (0.86) | 2.30 (0.50) | 0.81 (0.22) | 0.43 (0.11) | 0.38 (0.10) | 0 |
| 33 | Aklasis ežeras | 55.135, 24.482 | 16 | 3.50 (0.87) | 2.06 (0.30) | 0.79 (0.19) | 0.44 (0.10) | 0.50 (0.12) | 0 |
| 34 | Zapyškis forest | 54.898, 23.613 | 16 | 3.88 (0.67) | 2.15 (0.30) | 0.87 (0.14) | 0.49 (0.07) | 0.53 (0.10) | 1 |
| 35 | Aukštumala bog | 55.380, 21.367 | 15 | 3.50 (0.54) | 2.05 (0.26) | 0.82 (0.14) | 0.47 (0.08) | 0.43 (0.10) | 2 |
| 36 | Novaraistis | 54.959, 23.373 | 16 | 3.50 (0.73) | 2.42 (0.40) | 0.91 (0.17) | 0.52 (0.08) | 0.52 (0.08) | 0 |
| 37 | Labanoras forest | 55.217, 25.662 | 16 | 3.75 (0.86) | 2.23 (0.39) | 0.82 (0.20) | 0.45 (0.11) | 0.44 (0.11) | 0 |
| 38 | Viešvile NR | 55.176, 22.468 | 16 | 3.75 (0.94) | 2.24 (0.34) | 0.85 (0.19) | 0.48 (0.09) | 0.43 (0.11) | 0 |
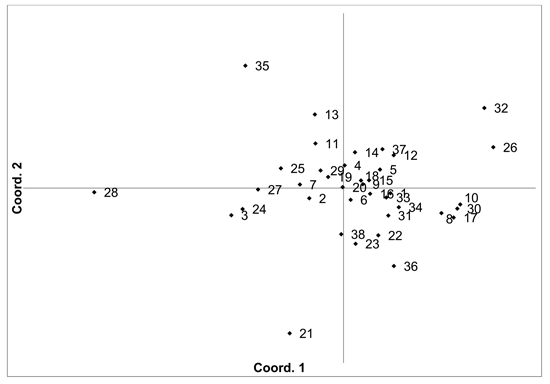
Fig. 2. Plot of Coordinate 1 vs. Coordinate 2 obtained by principal coordinates analysis (PCoA) based on pairwise unbiased Nei genetic distances between Vaccinium myrtillus populations. Population numbers as in Table 2. Variation explained by axis 1: 31.64%, axis 2: 22.82%.
| Table 3. Genetic diversity parameters, obtained using EST-SSR markers in the analysed Vaccinum vitis-idaea populations. | |||||||||
| Popula- tion number | Population | Coordinates (N, E) | No. of indivi- duals | Average no. of alleles (SE) | Average no. of effective alles (SE) | Average Information index (SE) | Average Unbiased Expected heterozy-gosity (SE) | Average Observed heterozy- gosity (SE) | No. of unique frag- ments |
| Latvia | |||||||||
| 1 | Preiļi | 56.368, 26.771 | 26 | 5.00 (0.67) | 3.27 (0.35) | 1.27 (0.12) | 0.67 (0.04) | 0.518 (0.050) | 0 |
| 2 | Rēzekne | 56.434, 27.378 | 24 | 5.20 (0.73) | 3.09 (0.48) | 1.24 (0.14) | 0.64 (0.04) | 0.575 (0.082) | 0 |
| 3 | Līvāni | 56.386, 26.235 | 24 | 4.50 (0.52) | 2.74 (0.29) | 1.10 (0.11) | 0.61 (0.05) | 0.508 (0.093) | 0 |
| 4 | Daugavpils | 55.956, 26.497 | 26 | 3.70 (0.45) | 2.56 (0.35) | 0.97 (0.15) | 0.53 (0.08) | 0.446 (0.119) | 0 |
| 5 | Embūte | 56.461, 21.887 | 24 | 5.30 (0.65) | 3.59 (0.36) | 1.36 (0.11) | 0.71 (0.03) | 0.562 (0.074) | 0 |
| 6 | Brocēni | 56.654, 22.638 | 24 | 5.30 (0.76) | 3.45 (0.39) | 1.31 (0.14) | 0.68 (0.05) | 0.487 (0.087) | 0 |
| 7 | Saka | 56.869, 21.304 | 25 | 5.80 (1.04) | 3.72 (0.63) | 1.33 (0.17) | 0.68 (0.05) | 0.531 (0.076) | 0 |
| 8 | Īvande | 57.004, 21.792 | 24 | 3.90 (0.55) | 2.35 (0.27) | 0.93 (0.14) | 0.51 (0.07) | 0.421 (0.095) | 0 |
| 9 | Ovīši | 57.564, 21.797 | 24 | 4.70 (0.70) | 2.81 (0.29) | 1.14 (0.13) | 0.61 (0.05) | 0.491 (0.093) | 0 |
| 10 | Gulbene | 57.217, 26.594 | 24 | 4.30 (0.58) | 2.35 (0.21) | 0.97 (0.11) | 0.54 (0.06) | 0.638 (0.113) | 0 |
| 11 | Alūksne | 57.410, 27.275 | 24 | 5.20 (0.63) | 3.18 (0.27) | 1.27 (0.11) | 0.67 (0.04) | 0.531 (0.080) | 0 |
| 12 | Beberbeķu parks | 56.952, 23.957 | 24 | 5.80 (0.74) | 4.05 (0.55) | 1.43 (0.16) | 0.70 (0.06) | 0.571 (0.069) | 2 |
| 13 | Zalvīte | 56.372, 25.208 | 24 | 5.30 (0.75) | 3.53 (0.51) | 1.31 (0.14) | 0.68 (0.05) | 0.616 (0.078) | 0 |
| 14 | Neveja | 57.564, 22.307 | 24 | 5.10 (0.71) | 3.64 (0.48) | 1.34 (0.13) | 0.70 (0.04) | 0.631 (0.058) | 0 |
| 15 | Ugāle | 57.289, 22.047 | 24 | 6.00 (0.70) | 3.39 (0.52) | 1.31 (0.13) | 0.67 (0.04) | 0.516 (0.089) | 2 |
| 16 | Rucava | 56.195, 21.107 | 24 | 4.60 (0.60) | 3.15 (0.44) | 1.20 (0.14) | 0.64 (0.05) | 0.532 (0.080) | 2 |
| 17 | Garciems | 57.110, 24.193 | 24 | 4.30 (0.56) | 2.64 (0.28) | 1.09 (0.12) | 0.59 (0.05) | 0.461 (0.093) | 0 |
| 18 | Oleri | 57.802, 25.414 | 24 | 5.10 (0.80) | 3.28 (0.36) | 1.24 (0.15) | 0.65 (0.06) | 0.504 (0.078) | 0 |
| 19 | Lubāna | 56.892, 26.682 | 26 | 4.90 (0.74) | 3.11 (0.36) | 1.22 (0.12) | 0.65 (0.04) | 0.485 (0.068) | 1 |
| 20 | Jumurda | 56.955, 25.725 | 24 | 5.20 (0.57) | 3.22 (0.29) | 1.29 (0.10) | 0.68 (0.03) | 0.600 (0.050) | 0 |
| Estonia | |||||||||
| 21 | Nigula | 58.007, 24.714 | 15 | 4.10 (0.53) | 2.70 (0.25) | 1.11 (0.10) | 0.62 (0.04) | 0.534 (0.086) | 0 |
| 22 | Rannametsa | 58.134, 24.512 | 16 | 5.10 (0.81) | 3.59 (0.48) | 1.32 (0.15) | 0.70 (0.04) | 0.608 (0.097) | 0 |
| 23 | Mustvee | 58.870, 26.919 | 16 | 4.70 (0.68) | 3.47 (0.54) | 1.27 (0.14) | 0.68 (0.05) | 0.620 (0.086) | 0 |
| 24 | Korvemaa | 59.285, 25.611 | 16 | 4.80 (0.76) | 3.48 (0.53) | 1.26 (0.15) | 0.67 (0.05) | 0.527 (0.083) | 0 |
| 25 | Peedu | 58.248, 26.453 | 16 | 5.00 (0.83) | 3.34 (0.58) | 1.22 (0.18) | 0.63 (0.07) | 0.478 (0.106) | 1 |
| 26 | Lehtmetsa | 59.203, 25.600 | 16 | 5.00 (0.70) | 3.32 (0.44) | 1.26 (0.15) | 0.66 (0.06) | 0.390 (0.075) | 0 |
| 27 | Tooma | 58.876, 26.272 | 16 | 4.40 (0.54) | 2.91 (0.22) | 1.17 (0.09) | 0.66 (0.03) | 0.544 (0.083) | 0 |
| Lithuania | |||||||||
| 28 | Motiejunai village | 55.051, 24.485 | 16 | 4.60 (0.73) | 3.09 (0.41) | 1.17 (0.16) | 0.63 (0.06) | 0.622 (0.085) | 1 |
| 29 | Degutine forest | 55.315, 25.738 | 16 | 4.70 (0.65) | 3.44 (0.50) | 1.25 (0.15) | 0.66 (0.06) | 0.580 (0.090) | 0 |
| 30 | Anykščiai | 55.566, 25.126 | 16 | 5.70 (0.84) | 3.44 (0.40) | 1.34 (0.14) | 0.68 (0.05) | 0.478 (0.081) | 0 |
| 31 | Zapyškis forest | 54.898, 23.613 | 15 | 4.80 (0.65) | 3.19 (0.38) | 1.24 (0.13) | 0.66 (0.05) | 0.556 (0.078) | 0 |
| 32 | Aklasis ežeras | 55.135, 24.482 | 16 | 5.10 (0.67) | 3.30 (0.46) | 1.26 (0.14) | 0.66 (0.05) | 0.466 (0.102) | 2 |
| 33 | Novaraistis | 54.959, 23.373 | 16 | 4.40 (0.43) | 2.91 (0.24) | 1.18 (0.09) | 0.66 (0.04) | 0.557 (0.071) | 1 |
| 34 | Labanoras forest | 55.217, 25.662 | 16 | 4.80 (0.53) | 3.21 (0.41) | 1.24 (0.12) | 0.66 (0.05) | 0.506 (0.086) | 0 |
| 35 | Viešvile NR | 55.176, 22.468 | 16 | 4.80 (0.59) | 3.47 (0.33) | 1.32 (0.11) | 0.71 (0.03) | 0.600 (0.084) | 1 |
| 36 | Aukštumala bog | 55.380, 21.367 | 16 | 4.60 (0.43) | 3.31 (0.38) | 1.25 (0.12) | 0.67 (0.05) | 0.479 (0.087) | 0 |
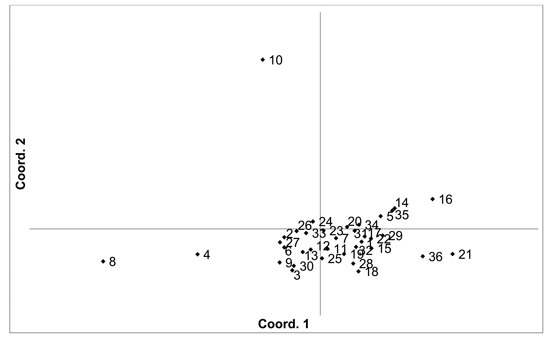
Fig. 3. Plot of Coordinate 1 vs. Coordinate 2 obtained by principal coordinates analysis (PCoA) based on of pairwise unbiased Nei genetic distances between Vaccinum vitis-idaea populations. Population numbers as in Table 3. Variation explained by axis 1: 23.89%, axis 2: 20.34%.
| Table 4. Genetic diversity parameters obtained using EST-SSR markers in the analysed Vaccinum uliginosum populations. | ||||||||
| Population number | Population | Coordinates (N, E) | No. of individuals | Total no. of fragments | No. of polymorphic fragments | Average Unbiased Expected heterozygosity (SE) | Average PIC (SE) | No. of unique fragments |
| Latvia | ||||||||
| 1 | Sala | 56.524, 25.727 | 24 | 110 | 109 | 0.117 (0.011) | 0.247 (0.016) | 0 |
| 2 | Zalvīte | 56.372, 25.208 | 26 | 96 | 95 | 0.116 (0.012) | 0.264 (0.018) | 0 |
| 3 | Kurmene | 56.427, 24.817 | 24 | 100 | 99 | 0.111 (0.011) | 0.246 (0.016) | 0 |
| 4 | Inčukalns | 57.084, 24.676 | 42 | 108 | 107 | 0.115 (0.011) | 0.246 (0.017) | 0 |
| 5 | Līvāni | 56.415, 26.282 | 48 | 124 | 124 | 0.12 (0.011) | 0.219 (0.015) | 0 |
| 6 | Saka | 56.869, 21.302 | 20 | 93 | 93 | 0.116 (0.011) | 0.275 (0.018) | 0 |
| 7 | Embūte | 56.461, 21.887 | 21 | 86 | 85 | 0.106 (0.011) | 0.275 (0.019) | 0 |
| 8 | Īvande | 57.005, 21.795 | 24 | 89 | 88 | 0.115 (0.012) | 0.287 (0.019) | 0 |
| 9 | Melnsils | 57.540, 22.740 | 22 | 90 | 89 | 0.108 (0.011) | 0.269 (0.019) | 0 |
| 10 | Viļaka | 57.155, 27.537 | 22 | 97 | 97 | 0.117 (0.011) | 0.275 (0.018) | 1 |
| 11 | Alūksne | 57.404, 27.269 | 23 | 69 | 68 | 0.104 (0.013) | 0.293 (0.022) | 0 |
| 12 | Gulbene | 57.217, 26.594 | 24 | 91 | 90 | 0.109 (0.011) | 0.266 (0.019) | 0 |
| 13 | Ugāle | 57.250, 22.033 | 23 | 99 | 98 | 0.107 (0.011) | 0.249 (0.017) | 1 |
| 14 | Brocēni | 56.656, 22.675 | 22 | 100 | 100 | 0.118 (0.012) | 0.256 (0.017) | 2 |
| 15 | Kalnciems | 56.803, 23.478 | 24 | 94 | 93 | 0.108 (0.011) | 0.257 (0.018) | 0 |
| 16 | Ķegums | 56.691, 24.669 | 24 | 87 | 87 | 0.109 (0.012) | 0.268 (0.018) | 0 |
| 17 | Beberbeķu parks | 56.952, 23.957 | 24 | 91 | 90 | 0.105 (0.011) | 0.264 (0.018) | 0 |
| 18 | Daugavas dzirnavas | 56.609, 24.891 | 14 | 78 | 77 | 0.112 (0.012) | 0.308 (0.021) | 0 |
| 19 | Neveja | 57.564, 22.307 | 56 | 114 | 113 | 0.11 (0.011) | 0.228 (0.016) | 0 |
| 20 | Oleri | 57.797, 25.473 | 46 | 106 | 105 | 0.11 (0.011) | 0.234 (0.017) | 0 |
| 21 | Jumurda | 56.975, 25.723 | 26 | 95 | 94 | 0.106 (0.011) | 0.258 (0.018) | 1 |
| 22 | Lubāna | 56.900, 26.762 | 41 | 107 | 106 | 0.114 (0.011) | 0.244 (0.017) | 0 |
| Estonia | ||||||||
| 23 | Korvemaa | 59.285, 25.611 | 16 | 86 | 85 | 0.115 (0.012) | 0.291 (0.019) | 0 |
| 24 | Tooma | 58.876, 26.272 | 15 | 96 | 95 | 0.116 (0.011) | 0.281 (0.018) | 1 |
| 25 | Lehtmetsa | 59.203, 25.600 | 15 | 79 | 78 | 0.115 (0.012) | 0.310 (0.021) | 0 |
| 26 | Nigula | 58.007, 24.714 | 16 | 93 | 92 | 0.115 (0.011) | 0.279 (0.018) | 1 |
| 27 | Rannametsa | 58.134, 24.512 | 13 | 77 | 76 | 0.108 (0.012) | 0.306 (0.021) | 0 |
| Lithuania | ||||||||
| 28 | Motiejunai | 55.051, 24.485 | 11 | 65 | 63 | 0.102 (0.013) | 0.316 (0.022) | 1 |
| 29 | Degutine forest | 55.315, 25.738 | 15 | 90 | 89 | 0.112 (0.011) | 0.273 (0.018) | 0 |
| 30 | Zapyškis forest | 54.898, 23.613 | 10 | 73 | 72 | 0.115 (0.013) | 0.320 (0.021) | 0 |
| 31 | Romuldava forest | 55.598, 25.213 | 15 | 83 | 82 | 0.112 (0.012) | 0.282 (0.019) | 1 |
| 32 | Labanoras forest | 55.217, 25.662 | 14 | 75 | 74 | 0.109 (0.012) | 0.304 (0.021) | 0 |
| 33 | Novaraistis | 54.959, 23.373 | 16 | 66 | 65 | 0.092 (0.011) | 0.294 (0.021) | 0 |
| 34 | Viešvile NR | 55.176, 22.468 | 14 | 79 | 78 | 0.117 (0.013) | 0.305 (0.021) | 0 |
| 35 | Aukštumala bog | 55.380, 21.367 | 16 | 87 | 86 | 0.113 (0.011) | 0.286 (0.018) | 1 |
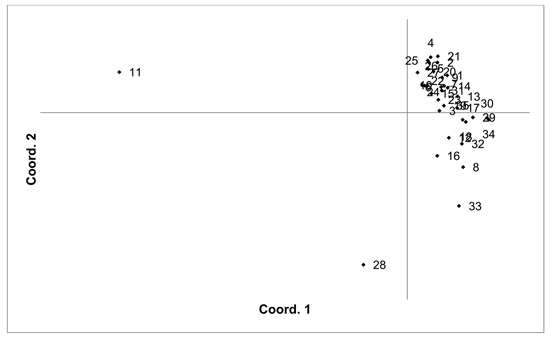
Fig. 4. Plot of Coordinate 1 vs. Coordinate 2 obtained by principal coordinates analysis (PCoA) based on pairwise unbiased Nei genetic distances between Vaccinum uliginosum populations. Population numbers as in Table 4. Variation explained by axis 1: 48.48%, axis 2: 14.20%.
| Table 5. Distribution (%) of chloroplast markers CP6 and ccmp7 alleles in Latvia, Estonia and Lithuania (%) in Vaccinium species | ||||||||||||
| V. myrtillus | V. vitis-idaea | V. uliginosum | ||||||||||
| marker | allele | Latvia | Estonia | Lithuania | allele | Latvia | Estonia | Lithuania | allele | Latvia | Estonia | Lithuania |
| n | 64 | 64 | 64 | 94 | 47 | 45 | ||||||
| CP6 | 153 | 29.7 | 0 | 7.8 | 158 | 38.3 | 29.8 | 24.4 | ||||
| 154 | 70.3 | 100 | 92.2 | 159 | 61.7 | 70.2 | 75.6 | |||||
| n | 169 | 28 | 36 | 103 | 52 | 82 | ||||||
| ccmp7 | 116 | 0.6 | 121 | 79.6 | 73.1 | 83.1 | ||||||
| 117 | 89.3 | 82.1 | 86.1 | 122 | 3.2 | |||||||
| 120 | 9.5 | 17.9 | 13.9 | 123 | 20.4 | 26.9 | 13.7 | |||||
| 121 | 0.6 | |||||||||||