| Table 1. RAPD primers used for fingerprinting bamboo species from a Brazilian collection. | |
| Identification | Sequence |
| OPC-20 | 5’-ACTTCGCCAC-3’ |
| OPV-17 | 5’-ACCGGCTTGT-3’ |
| OPG-17 | 5’-ACGACCGACA-3’ |
| OPAN-03 | 5’-AGCCAGGCTG-3’ |
| OPB-10 | 5’-CTGCTGGGAC-3’ |
| OPAW-20 | 5’-TGTCCTAGCC-3’ |
| OPAW-08 | 5’-CTGTCTGTGG-3’ |
| OPJ-01 | 5’-CCCGGCATAA-3’ |
| OPJ-15 | 5’-TGTAGCAGGG-3’ |
| OPJ-04 | 5’-CCGAACACGG-3’ |
| OPI-15 | 5’-TCATCCGAGG-3’ |
| OPL-04 | 5’-GACTGCACAC-3’ |
| Table 2. Total number of bands (N), species with unique bands (UB) and number of unique bands detected with RAPD and RAPD-RFLP markers in bamboo species. | ||||||||||||||||
| Marker | N | UB | PIC | Unique bands per species | ||||||||||||
| RAPD | BV | BVV | BB | DG | DA | PE | PH | GA | GS | G1 | G2 | G3 | G4 | |||
| OPC-20 | 1 | 0 | 0.00 | |||||||||||||
| OPV-17 | 5 | 1 | 0.74 | 1 | ||||||||||||
| OPG-17 | 7 | 2 | 0.85 | 1 | 1 | |||||||||||
| OPAN-03 | 7 | 1 | 0.71 | 1 | ||||||||||||
| OPB-10 | 10 | 2 | 0.78 | 1 | 1 | |||||||||||
| OPAW-20 | 9 | 4 | 0.91 | 1 | 1 | 1 | 1 | |||||||||
| OPAW-08 | 5 | 1 | 0.88 | 1 | ||||||||||||
| OPJ-01 | 5 | 3 | 0.94 | 1 | 2 | |||||||||||
| OPJ-15 | 6 | 1 | 0.56 | 1 | ||||||||||||
| OPJ-04 | 7 | 1 | 0.84 | 1 | ||||||||||||
| OPI-15 | 12 | 2 | 0.89 | 1 | 1 | |||||||||||
| OPL-04 | 9 | 1 | 0.81 | 1 | ||||||||||||
| Total | 83 | 19 | 0.74 | 1 | 2 | 4 | 2 | 0 | 1 | 5 | 3 | 0 | 1 | 0 | 0 | 0 |
| RAPD-RFLP | ||||||||||||||||
| MspI | 33 | 9 | 0.86 | 2 | 4 | 1 | 2 | |||||||||
| HindIII/HaeIII | 26 | 14 | 0.94 | 5 | 1 | 2 | 4 | 1 | 1 | |||||||
| HinfI/RsaI | 35 | 11 | 0.93 | 4 | 3 | 3 | 1 | |||||||||
| Total | 94 | 34 | 0.91 | 0 | 2 | 13 | 0 | 2 | 5 | 2 | 4 | 0 | 3 | 1 | 1 | 1 |
| BV – Bambusa vulgaris. BVV – Bambusa vulgaris vittata. BB – Bambusa beecheyana. DG – Dendrocalamus giganteus. DA – Dendrocalamus asper. PE – Phyllostachys edulis. PH – Phyllostachys heterocycla. GA – Guadua amplexifolia. GS – Guadua superba. G1, G2, G3 and G4 – Guadua spp. | ||||||||||||||||
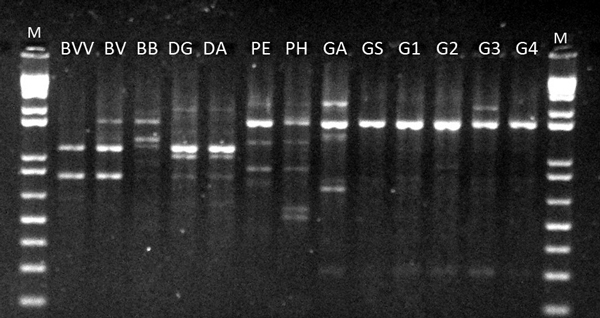
Fig. 1. RAPD profile of OPV-17 for 13 bamboo taxa. BVV – Bambusa vulgaris vittata. BV – Bambusa vulgaris. BB – Bambusa beecheyana. PE – Phyllostachys edulis. PH – Phyllostachys heterocycla. DA – Dendrocalamus asper. DG – Dendrocalamus giganteus. GA – Guadua amplexifolia. GS – Guadua superba. G1, G2, G3 and G4 – Guadua spp. M – 1 Kb Plus Ladder DNA.
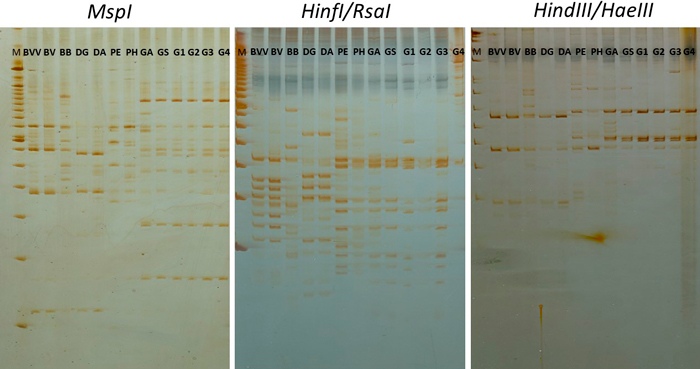
Fig. 2. RAPD-RFLP profile from the digestion of RAPD products from primer OPV17 with three restriction enzyme combinations. BVV – Bambusa vulgaris vittata. BV – Bambusa vulgaris. BB – Bambusa beecheyana. DG – Dendrocalamus giganteus. DA – Dendrocalamus asper. PE – Phyllostachys edulis. PH – Phyllostachys heterocycla. GA – Guadua amplexifolia. GS – Guadua superba. G1, G2, G3 and G4 – Guadua spp. M – 100 bp marker (Ladder).
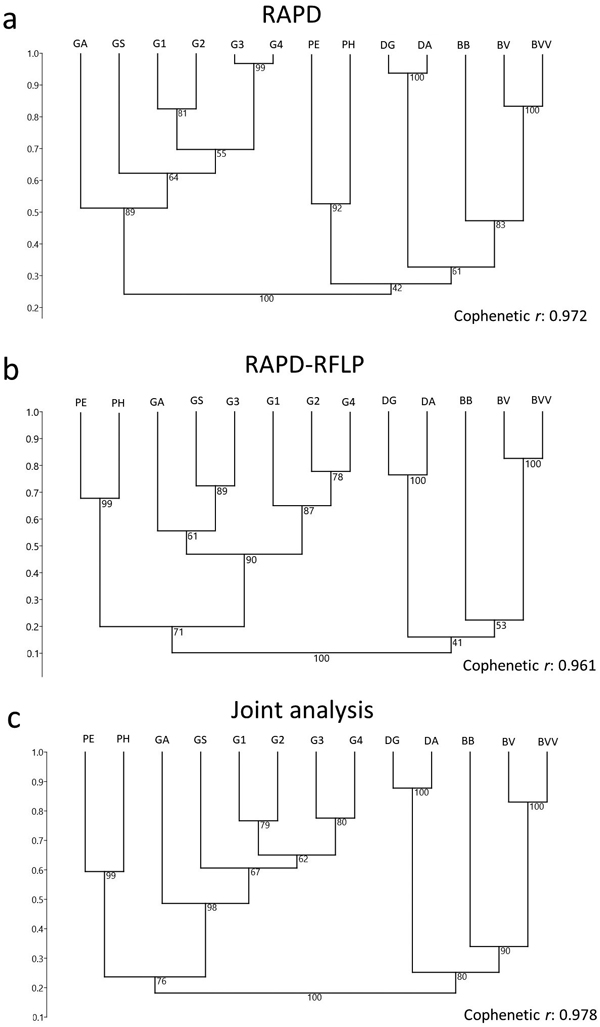
Fig. 3. Dendrogram of 13 bamboo taxa based on (a) RAPD and (b) RAPD-RFLP markers, and the (c) joint analysis. BVV – Bambusa vulgaris vittata. BV – Bambusa vulgaris. BB – Bambusa beecheyana. PE – Phyllostachys edulis. PH – Phyllostachys heterocycla. DA – Dendrocalamus asper. DG – Dendrocalamus giganteus. GA – Guadua amplexifolia. GS – Guadua superba. G1, G2, G3 and G4 – Guadua spp.
| Table 3. Jaccard’s similarity coefficient among 13 bamboo taxa, based on 83 RAPD markers. | |||||||||||||
| BV | BVV | BB | DG | DA | PE | PH | GA | GS | G1 | G2 | G3 | G4 | |
| BV | 0.833 | 0.486 | 0.304 | 0.318 | 0.325 | 0.271 | 0.212 | 0.204 | 0.245 | 0.235 | 0.18 | 0.184 | |
| BVV | 0.459 | 0.283 | 0.295 | 0.3 | 0.25 | 0.216 | 0.184 | 0.226 | 0.216 | 0.184 | 0.188 | ||
| BB | 0.372 | 0.39 | 0.3 | 0.304 | 0.265 | 0.234 | 0.226 | 0.216 | 0.208 | 0.213 | |||
| DG | 0.938 | 0.213 | 0.275 | 0.264 | 0.286 | 0.296 | 0.264 | 0.212 | 0.216 | ||||
| DA | 0.222 | 0.286 | 0.275 | 0.298 | 0.308 | 0.275 | 0.22 | 0.224 | |||||
| PE | 0.526 | 0.333 | 0.244 | 0.189 | 0.2 | 0.217 | 0.222 | ||||||
| PH | 0.447 | 0.28 | 0.268 | 0.259 | 0.255 | 0.26 | |||||||
| GA | 0.61 | 0.521 | 0.489 | 0.467 | 0.477 | ||||||||
| GS | 0.683 | 0.692 | 0.55 | 0.564 | |||||||||
| G1 | 0.825 | 0.683 | 0.659 | ||||||||||
| G2 | 0.737 | 0.711 | |||||||||||
| G3 | 0.968 | ||||||||||||
| BV – Bambusa vulgaris. BVV – Bambusa vulgaris vittata. BB – Bambusa beecheyana. DG – Dendrocalamus giganteus. DA – Dendrocalamus asper. PE – Phyllostachys edulis. PH – Phyllostachys heterocycla. GA – Guadua amplexifolia. GS – Guadua superba. G1, G2, G3 and G4 – Guadua spp. | |||||||||||||
| Table 4. Jaccard’s similarity coefficient among 13 bamboo taxa, based on 94 RAPD-RFLP markers. | |||||||||||||
| BBV | BV | BB | DG | DA | PE | PH | GA | GS | G1 | G2 | G3 | G4 | |
| BBV | 0.826 | 0.209 | 0.172 | 0.188 | 0.114 | 0.154 | 0.106 | 0.098 | 0.086 | 0.121 | 0.093 | 0.121 | |
| BV | 0.238 | 0.172 | 0.188 | 0.14 | 0.154 | 0.13 | 0.098 | 0.086 | 0.121 | 0.119 | 0.121 | ||
| BB | 0.1 | 0.143 | 0.157 | 0.146 | 0.192 | 0.196 | 0.116 | 0.146 | 0.213 | 0.146 | |||
| DG | 0.765 | 0.025 | 0.028 | 0.023 | 0.057 | 0 | 0.036 | 0.054 | 0.036 | ||||
| DA | 0.047 | 0.051 | 0.067 | 0.108 | 0.062 | 0.1 | 0.103 | 0.1 | |||||
| PE | 0.677 | 0.311 | 0.3 | 0.098 | 0.128 | 0.227 | 0.1 | ||||||
| PH | 0.341 | 0.333 | 0.108 | 0.111 | 0.22 | 0.111 | |||||||
| GA | 0.571 | 0.371 | 0.382 | 0.541 | 0.382 | ||||||||
| GS | 0.519 | 0.538 | 0.724 | 0.538 | |||||||||
| G1 | 0.65 | 0.433 | 0.65 | ||||||||||
| G2 | 0.5 | 0.778 | |||||||||||
| G3 | 0.556 | ||||||||||||
| BVV – Bambusa vulgaris vittata. BV – Bambusa vulgaris. BB – Bambusa beecheyana. DG – Dendrocalamus giganteus. DA – Dendrocalamus asper. PE – Phyllostachys edulis. PH – Phyllostachys heterocycla. GA – Guadua amplexifolia. GS – Guadua superba. G1, G2, G3 and G4 – Guadua spp. | |||||||||||||
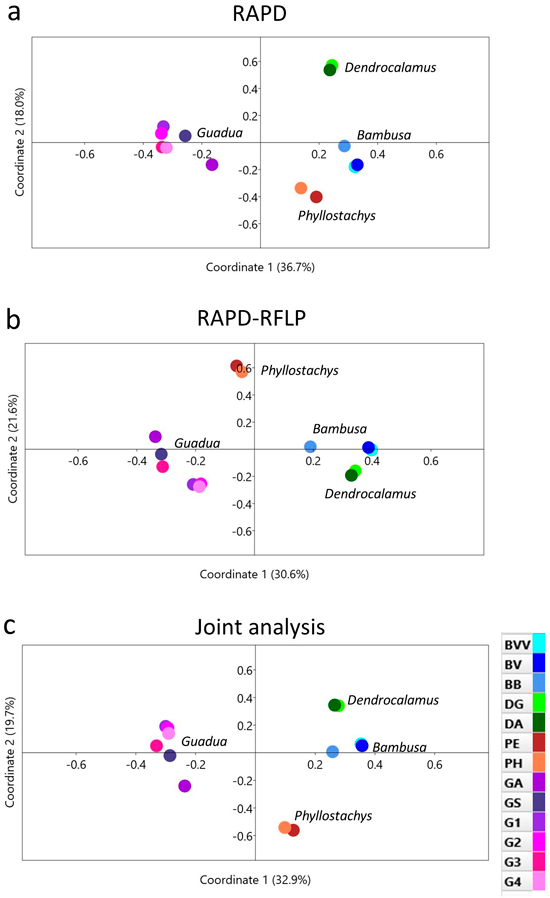
Fig. 4. Principal coordinate analysis of 13 bamboo taxa based on (a) RAPD, (b) RAPD-RFLP and (c) the joint analysis. BVV – Bambusa vulgaris vittata. BV – Bambusa vulgaris. BB – Bambusa beecheyana. DG – Dendrocalamus giganteus. DA – Dendrocalamus asper. PE – Phyllostachys edulis. PH – Phyllostachys heterocycla. GA – Guadua amplexifolia. GS – Guadua superba. G1, G2, G3 and G4 – Guadua spp.