| Table 1. The geographical characteristics of four populations of C. sativa in Hyrcanian forest and the sample size in each population. | ||||
| Population name | Latitude, Longitude | Altitude (m a.s.l.) | Sample size (No.) | Population size (Hectare) |
| Veysroud (V) | 37°16´N, 49°15´E | 211–711 | 8 | 60 |
| Shafaroud (SH) | 37°30´N, 49°02´E | 200–360 | 8 | 50 |
| Siyahmazgi (S) | 40°97´N, 35°49´E | 290–350 | 8 | 40 |
| QaleRoudkhan (R) | 37°05´N, 49°14´E | 200–400 | 8 | 50 |
Fig. 1. The geographical positions of Hyrcanian populations of C. sativa in North of Iran. R = QalehRoudkhan region; S = SiyahMazgi region; SH = Shafaroud region; V = Veysroud region.
| Table 2. Sequences of Castanea SSR primer pairs, PCR annealing temperature and PCR expected product size. | |||||||
| Marker | Number | SSR locus | Primer sequences (5’ → 3’) | Ta (°C) | Size of alleles (bp) | Reference | |
| NU SSR | 1 | CsCAT 1 | F | GAGAATGCCCACTTTTGCA | 54 | 177–212 | Marinoni et al. 2003 |
| R | GCTCCCTTATGGTCTCG | ||||||
| 2 | CsCAT 3 | F | CACTATTTTATCATGGACGG | 58 | 179–198 | ||
| R | CGAATTGAGAGTTCATACTC | ||||||
| 3 | CsCAT 6 | F | AGTGCTCGTGGTCAGTGAG | 54 | 199–238 | ||
| R | CAACTCTGCATGATAAC | ||||||
| 4 | CsCAT 7 | F | GAACATGATGATTGGCCTC | 54 | 179–223 | ||
| R | CCAAACATGACATATGTCCC | ||||||
| 5 | CsCAT 14 | F | CGAGGTTGTTGTTCATCATTAC | 54 | 133–151 | ||
| R | GATCTCAAGTCAAAAGGTGTC | ||||||
| 6 | CsCAT 15 | F | TTCTGCGACCTCGAAACCGA | 56 | 123–155 | ||
| R | GCTAGGGTTTTCATTTCTAG | ||||||
| 7 | CsCAT 17 | F | TTGGCTATACTTGTTCTGCAAG | 56 | 132–151 | ||
| R | GCCCCATGTTTTCTTCCATGG | ||||||
| 8 | CsCAT 41 | F | AAGTCAGCAACACCATATGC | 58 | 199–238 | ||
| R | CCCACTGTTCATGAGTTTCT | ||||||
| 9 | EMC 38 | F | TTTCCCTATTTCTAGTTTGTGATG | 58 | 227–261 | Buck et al. 2003 | |
| R | ATGGCGCTTTGGATGAAC | ||||||
| 10 | QpZAG110 | F | GGAGGCTTCCTTCAACCTACT | 54 | 207–223 | Steinkellner et al. 1997 | |
| R | GATCTCTTGTGTGCTGTATTT | ||||||
| CP SSR | 1 | CCMP2 | F | GATCCCGGACGTAATCCTG | 55 | 234 | Weising and Gardner 1999 |
| R | ATCGTACCGAGGGTTCGAAT | ||||||
| 2 | CCMP4 | F | AATGCTGAATCGAYGACCTA | 55 | 114 | ||
| R | CCAAAATATTBGGAGGACTCT | ||||||
| 3 | CCMP10 | F | TTTTTTTTTAGTGAACGTGTCA | 55 | 113 | ||
| R | TTCGTCGDCGTAGTAAATAG | ||||||
| 4 | CMCS2 | F | GAGCCATTCCCTTTTAGAAT | 55 | 141 | Sebastiani et al. 2004 | |
| R | TTGAAAACCGGTATAGTTCG | ||||||
| 5 | CMCS7 | F | AAGCGAGATGAATGAGTTTT | 55 | 205 | ||
| R | AAAATTGGATTGATTATTGACT | ||||||
| 6 | CMCS8 | F | GGTCTATTTTTCCACTCACAA | 55 | 179 | ||
| R | AGAAATAAACACCCCCATTA | ||||||
| 7 | CMCS12 | F | ATATTGGTAAAACGGCAACT | 55 | 216 | ||
| R | TTTATGGCATGAAACAACTC | ||||||
| 8 | CMCS14 | F | GGATTGTAACAAATTTTTCAGG | 55 | 176 | ||
| R | GTGCAAGGAATGTCGAACTA | ||||||
| F = forward primer; R = reverse primer; SSR = Simple sequence repeat; CP SSR = Chloroplast SSR; NU SSR = Nuclear SSR; Ta = PCR annealing temperature | |||||||
| Table 3. Genetic variability within four C. sativa populations based on SSR markers. | ||||||||||
| Marker | Pop | N | Na | Ne | I | Ho | He | UHe | F | |
| NU SSR | R | Mean | 8.000 | 3.000 | 2.142 | 0.807 | 0.538 | 0.463 | 0.494 | –0.147 |
| SE | 0.000 | 0.365 | 0.243 | 0.133 | 0.110 | 0.074 | 0.078 | 0.125 | ||
| S | Mean | 8.000 | 2.400 | 1.903 | 0.679 | 0.563 | 0.434 | 0.463 | –0.293 | |
| SE | 0.000 | 0.221 | 0.148 | 0.093 | 0.112 | 0.059 | 0.063 | 0.166 | ||
| SH | Mean | 8.000 | 3.300 | 2.435 | 0.962 | 0.675 | 0.559 | 0.597 | –0.230 | |
| SE | 0.000 | 0.213 | 0.197 | 0.081 | 0.075 | 0.044 | 0.046 | 0.130 | ||
| V | Mean | 8.000 | 2.700 | 2.174 | 0.781 | 0.475 | 0.477 | 0.508 | –0.007 | |
| SE | 0.000 | 0.260 | 0.223 | 0.122 | 0.096 | 0.073 | 0.078 | 0.115 | ||
| Total | Mean | 8.000 | 2.85 | 2.163 | 0.630 | 0.563 | 0.512 | 0.516 | –0.169 | |
| SE | 0.000 | 0.265 | 0.203 | 0.107 | 0.098 | 0.063 | 0.071 | 0.134 | ||
| NU SSR = Nuclear SSR; R = QalehRoudkhan region; S = SiyahMazgi region; SH = Shafaroud region; V = Veysroud region; N = number of individuals analyzed; Na = number of alleles; Ne = effective number of alleles; I = Shannon index; Ho = observed heterozygosity; He = expected heterozygosity; UHe = unbiased expected heterozygosity; F = F-values; SE = Standard errors | ||||||||||
| Table 4. Analyses of molecular variance (AMOVA) for four populations of C. sativa from Hyrcanian forest by Nuclear SSR (NU SSR). Statistics include sums of squared deviations (SS); mean squared deviations (MS), variance component estimates (Est. Var.), the percentage of the total variance contributed by each component; estimator of relative genetic differentiation based on fraction of total variance of allele size between two subpopulations (Rst) and the probability of obtaining a more extreme component estimate by chance alone. | ||||||
| Marker | S.O.V. | D.f | SS | MS | Est. Var. | % of Variance |
| NU SSR | Between groups | 3 | 8.305.094 | 2.768.365 | 130.923 | 16% |
| Within groups | 60 | 40.415.375 | 673.590 | 673.590 | 84% | |
| Total Sum | 63 | 48.720.469 | 804.513 | 100% | ||
| Stat | Value(Φ) | P(rand > = data) | ||||
| Rst | 0.163 | 0.010 | ||||
| Table 5. Pairwise estimated of Nei’s genetic distance and the calculated Fst based on 10 nuclear SSR markers among four populations of C. sativa in Hyrcanian forest. | ||||
| Population | R | S | SH | |
| Nei distance | R | 0.000 | ||
| S | 0.215 | |||
| SH | 0.237 | 0.175 | 0.000 | |
| V | 0.012 | 0.095 | 0.208 | |
| Fst | R | 0.000 | ||
| S | 0.132 | 0.000 | ||
| SH | 0.185 | 0.148 | 0.000 | |
| V | 0.034 | 0.114 | 0.163 | |
| NU SSR = Nuclear SSR; R = QalehRoudkhan region; S = SiyahMazgi region; SH = Shafaroud region; V = Veysroud region; Fst = Fixation index | ||||
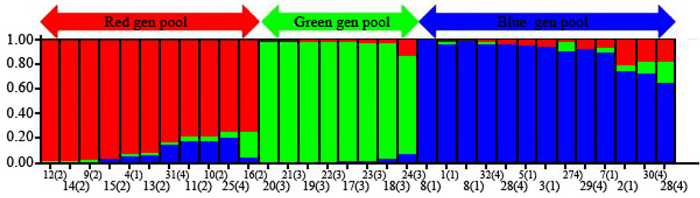
Fig. 2. Population structure of C. sativa was estimated based on Structure analysis. In the figure, the individuals of Iranian Castanea were sorted by Bayesian clustering approaches based on 10 nuclear microsatellite loci. Each bar represents a single individual analyzed. Legend of number in brackets: 1 = R1; 2 = R2; 3 = R3; 4 = R4; 5 = R5; 6 = R6; 7 = R7; 8 = R8; 9 = S1; 10 = S2; 11 = S3; 12 = S4; 13 = S5; 14 = S6; 15 = S7; 16 = S8; 17 = SH1; 18 = SH2; 19 = SH3; 20 = SH4; 21 = SH5; 22 = SH7; 23 = SH10; 24 = SH12; 25 = V1; 26 = V2; 27 = V3; 28 = V4; 29 = V5; 30 = V6; 31 = V7; 32 = V8.
| Table 6. The proportion membership of each individual of the four populations in each of the 3 gene pools identified by Structure analysis. | ||||||||||
| Sample | Structuregene pools (% inferred ancestry) | Sample | Structuregene pools (% inferred ancestry) | |||||||
| red | green | blue | red | green | blue | |||||
| 1 | R1 | 0.972 | 17 | SH1 | 0.950 | |||||
| 2 | R2 | 0.750 | 18 | SH2 | 0.930 | |||||
| 3 | R3 | 0.921 | 19 | SH3 | 0.972 | |||||
| 4 | R4 | 0.927 | 20 | SH4 | 0.976 | |||||
| 5 | R5 | 0.951 | 21 | SH5 | 0.974 | |||||
| 6 | R6 | 0.977 | 22 | SH7 | 0.967 | |||||
| 7 | R7 | 0.896 | 23 | SH10 | 0.943 | |||||
| 8 | R8 | 0.973 | 24 | SH12 | 0.803 | |||||
| 9 | S1 | 0.965 | 25 | V1 | 0.747 | |||||
| 10 | S2 | 0.784 | 26 | V2 | 0.649 | |||||
| 11 | S3 | 0.781 | 27 | V3 | 0.911 | |||||
| 12 | S4 | 0.978 | 28 | V4 | 0.956 | |||||
| 13 | S5 | 0.912 | 28 | V5 | 0.910 | |||||
| 14 | S6 | 0.973 | 30 | V6 | 0.727 | |||||
| 15 | S7 | 0.952 | 31 | V7 | 0.820 | |||||
| 16 | S8 | 0.744 | 32 | V8 | 0.964 | |||||
| R = QalehRoudkhan region; S = SiyahMazgi region; SH = Shafaroud region; V = Veysroud region | ||||||||||
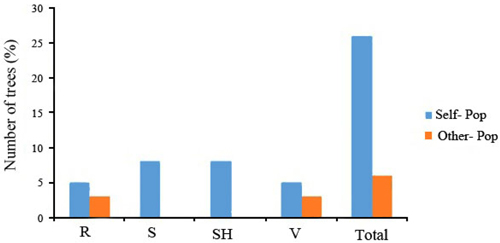
Fig. 3. Assignment percentage of individuals based on structure analysis per each population (K = 3). R = QalehRoudkhan region; S = SiyahMazgi region; SH = Shafaroud region; V = Veysroud region.
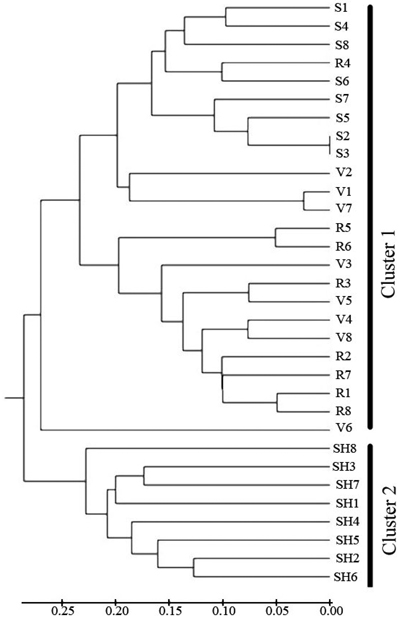
Fig. 4. Dendrogram depicting the distribution of genotypes of the four populations of C. sativa in Hyrcanian forest based on UMGMA method. R = QalehRoudkhan region; S = SiyahMazgi region; SH = Shafaroud region; V = Veysroud region.
| Table 7. Results of BOTTLENECK tests elaborated on the four C. sativa populations analysed at 10 polymorphicnuclear microsatellite loci. | ||||||
| Test | Mutation Model | Population | ||||
| V | SH | S | R | |||
| Sign | P-value | IAM | 0.08 | 0.225 | 0.272 | 0.347 |
| SMM | 0.034* | 0.807 | 0.513 | 0.359 | ||
| TPM | 0.072 | 0.359 | 0.335 | 0.369 | ||
| Wilcoxon | P-value | IAM | 0.359 | 0.921 | 0.496 | 1.000 |
| SMM | 0.203 | 0.625 | 0.734 | 0.82 | ||
| TPM | 0.250 | 0.625 | 0.57 | 0.91 | ||
| Standardized differences | P-value | IAM | <0.001* | 0.468 | 0.003* | 0.396 |
| SMM | <0.001* | 0.272 | 0.04* | 0.057 | ||
| TPM | <0.001* | 0.114 | 0.012 | 0.208 | ||
| * Significant at level 0.05; P = Probability; IAM = Infinite allele model, TPM = Two phase model, SMM = Stepwise mutation model; R = QalehRoudkhan region; S = SiyahMazgi region; SH = Shafaroud region; V = Veysroud region | ||||||