Genetic diversity and sexual reproduction in relict populations of Betula nana
Jadwiszczak K. A., Kłosowski S., Zalewska I., Banaszek A., Chrzanowska A. (2017). Genetic diversity and sexual reproduction in relict populations of Betula nana. Silva Fennica vol. 51 no. 2 article id 5643. https://doi.org/10.14214/sf.5643
Highlights
- Genetic diversity parameters and meiotic recombination frequencies in the relict populations were comparable to those from widespread localities
- Contribution of seeds without ovule was very high
- Fully developed seeds germinated better in central populations
- Significant differences of groundwater parameters were observed between relict and central populations.
Abstract
In the present study, the impact of geographical isolation and habitat conditions on genetic diversity and sexual reproduction was tested in four relict populations of dwarf birch Betula nana L. in Poland and Belarus. Amplified fragment length polymorphism (AFLP) method revealed that the endangered central European stands were not genetically extirpated compared with the widespread localities from Finland and Russia, which can result from infrequent outcrossing events in long-living clonal populations. However, genetic clustering methods indicated significant differentiation of the Polish populations because of their small sizes and long-term geographical isolation. Considerable numbers of empty seeds were observed in both relict and central locations, although fully developed seeds germinated better in widespread populations. Analysis of groundwater chemical parameters indicated that two relict populations were significantly different from the remaining samples with respect to pH, electrical conductivity and concentrations of phosphorus ions, which can also influence the efficiency of sexual reproduction. In the light of results obtained it seems that endangered B. nana localities are relatively stable.
Keywords
flowering;
AFLP;
dwarf birch;
meiotic recombination;
seed germination
-
Jadwiszczak,
Institute of Biology, University of Białystok, Ciołkowskiego 1J, 15-245 Białystok, Poland
 http://orcid.org/0000-0002-9345-8891
E-mail
jadwiszczak2010@gmail.com
http://orcid.org/0000-0002-9345-8891
E-mail
jadwiszczak2010@gmail.com

- Kłosowski, Department of Environment Protection and Modelling, The Jan Kochanowski University, Świętokrzyska 15, 25-406 Kielce, Poland E-mail stanislaw.klosowski@ujk.kielce.pl
- Zalewska, Faculty of Pharmacy, Medical University of Białystok, Mickiewicza 2a, 15-222 Białystok, Poland E-mail iwonazalewska1988@gmail.com
- Banaszek, Institute of Biology, University of Białystok, Ciołkowskiego 1J, 15-245 Białystok, Poland E-mail banaszek@uwb.edu.pl
- Chrzanowska, Institute of Biology, University of Białystok, Ciołkowskiego 1J, 15-245 Białystok, Poland E-mail maga.chrzanowska@gmail.com
Received 23 January 2017 Accepted 22 March 2017 Published 28 March 2017
Views 112300
Available at https://doi.org/10.14214/sf.5643 | Download PDF

1 Introduction
The present pattern of genetic diversity distribution within the boundaries of species is a consequence of range shifts forced by Quaternary climatic changes (Hewitt 2004; Hampe and Petit 2005). During glacial cycles, the ranges of tropical and temperate species were significantly changed by the decay of individuals in unfavourable habitats, altitudinal population dislocations and fragmentation of populations in refugia (Jolly et al. 1998; Hewitt 2004). In turn, arctic and boreal taxa were either pushed to lower latitudes or divided into small temperate or northern refugia (Stewart and Lister 2001; Pruett and Winker 2008). Rising temperatures enabled an expansion of thermophile species from south to north, whereas cold-tolerant taxa either spread from high latitude refugia or contracted their ranges. However, some populations of boreal and arctic species have remained in small, isolated enclaves of suitable environments at the retreating range limit. Such populations are known as climatic relicts (Hampe and Jump 2011). Relict populations, being out of the continuous range, could be subjected to differential pressures of natural selection, strong genetic drift and limited gene flow. These evolutionary forces can drive small isolated populations to either significant genetic differentiation and consequently to speciation (Ellstrand and Elam 1993) or to genetic erosion resulting in a decrease of population productivity, growth and stability (Hughes et al. 2008; Bolnick et al. 2011).
One of the climatic relict plants in central Europe is the dwarf birch Betula nana L. Because the occurrence of dwarf birch is related to low temperatures, it was more widespread during glacial periods of the Weichselian than it is today (Provan and Bennett 2008). The current continuous European range of B. nana spreads over Greenland Island, the northern part of Scotland, Fennoscandia and Siberia. The species also exists in isolated populations in Germany, Poland, the Czech Republic, Belarus and Lithuania, where it is recognised as endangered (EN category of the International Union for Conservation of Nature). This is a monoecious, self-incompatible, wind-pollinated and wind-dispersed shrub. In the continuous range, the species occupies moist, acidic and nutrient-poor localities. Relict populations of the dwarf birch inhabit open raised-bogs, where the main threat is expansion of forest plants (Kruszelnicki et al. 2014; Schwartz and Poschlod 2015). B. nana is found in three nature reserves in Poland: ”Linje” (population code – LIN) in northern part of the country as well as ”Torfowisko pod Zieleńcem” (ZIEL) and ”Torfowiska Doliny Izery” (IZER) in the Sudety Mts.
Analysis of chloroplast DNA (cpDNA) diversity has revealed that all Polish populations are fixed in respect of the most common birch haplotypes (Jadwiszczak et al. 2012). LIN and ZIEL reserves share the same haplotype, whereas the IZER reserve is characterised by another haplotype. Based on this result, it was hypothesised that either Poland was populated from distinct glacial refugia or haplotype fixation resulted from genetic drift acting strongly in relict populations (Jadwiszczak et al. 2012). The aim of the present work was to assess resources of genetic diversity in three Polish and one Belarusian relict populations of B. nana using the amplified fragment length polymorphism (AFLP) method to determine if they are genetically eroded compared with the central localities from Finland and Russia. We also tested the extent of genetic differentiation among populations located within and out of the continuous range. Because the level of genetic variation mainly results from ongoing contemporary processes in populations (Paun et al. 2008), the next question we asked was about the effectiveness of sexual reproduction of the dwarf birches in Poland and Finland. We also determined if Polish and Finnish localities of dwarf birch were differentiated with respect to chemical groundwater parameters because it has been proven that the investment of wetland and grassland plants into sexual reproduction is related to the amount of nitrogen and phosphorus in the ground (Fujita et al. 2014).
2 Materials and methods
2.1 Sample collection and chemical analyses of groundwater
Detailed studies of genetic variation and reproduction were carried out in three endangered Polish populations of B. nana located out of the continuous range and three abundant Finnish populations from the centre of species distribution. Moreover, to assess the genetic differentiation, one sample from Belarus and one from Russia were included in the genetic investigations (Table 1, Fig. 1). In total, 139 individuals were collected for genetic analyses. Flowering was explored in 66 individuals from Poland and 74 from Finland (Table 2). Finnish and Polish populations were also analysed for seed weight and germination success (100 specimens). In general, reproduction analyses concerned the same individuals for which AFLP genotyping was performed, although in some cases genotyping failed. All Polish and Finnish localities were visited twice in 2013: in the spring to collect fresh leaves for DNA isolation and to count the male and female flowers and in autumn to sample the seeds. To increase the probability of collection of genetically distinct individuals (genets), samples were distant from one another by at least 10 m. The leaf material for DNA isolation was dried in silica gel, seeds were collected in paper bags, and plant material was stored at room temperature.
| Table 1. Population names, their geographical locations, number of individuals sampled for genetic analyses, parameters of genetic diversities and inbreeding coefficients (F) for B. nana sampling sites studied. Ploc = number of polymorphic loci; Hj = Nei’s gene diversity. | ||||||||
| Country | Population | Code | Location | No of individuals | Ploc | Hj | F | |
| Latitude, Longitude | ||||||||
| 1. | Poland | Linje | LIN | 53°11´N, 18°18´E | 23 | 83 | 0.120 | 0.249 |
| 2. | Torfowisko pod Zieleńcem | ZIEL | 50°20´N, 16°25´E | 20 | 91 | 0.129 | 0.130 | |
| 3. | Torfowiska Doliny Izery | IZER | 50°51´N, 15°21´E | 20 | 102 | 0.136 | 0.060 | |
| 4. | Finland | Juttuvaara | JUT | 62°57´N, 30°22´E | 21 | 87 | 0.103 | 0.075 |
| 5. | Kaitaa | KAI | 63°14´N, 28°52´E | 20 | 87 | 0.114 | 0.402 | |
| 6. | Tuusniemi | TUS | 62°52´N, 28°24´E | 21 | 91 | 0.091 | 0.035 | |
| 7. | Belarus | Berezin’skij Zapovednik | BZ | 54°37´N, 28°21´E | 7 | 85 | 0.120 | 0.078 |
| 8. | Russia | Ural Mts. | UR | 63°03´N, 58°36´E | 7 | 89 | 0.115 | 0.057 |
Fig. 1. Locations of study populations. Population codes according to Table 1.
| Table 2. Parameters of habitat quality and sexual reproduction in the B. nana populations. Pop = populations codes according to Table 1; Hab = number of habitat plots; Habitat quality parameters: EC = electrical conductivity; Reproductive parameters: GS = number of germinated (without scarification) seeds per individual, GSC = number of seeds germinated after scarification, ES = number of empty seeds (without ovule), PF = number of partly filled seeds, IN = number of seeds infected by insects; # = median values. View in new window/tab. |
To conduct chemical analyses of groundwater [pH, electrical conductivity (EC; µSm cm–1), the concentrations of NH4+ (mg l–1) and PO43– (mg l–1) ions], 5 to 10 habitat plots were set up during the spring and autumn at each locality in Poland and Finland (Table 2). In the field, EC and pH were determined using a CPC-401 pH-meter (Elmetron). In the laboratory, the concentrations of NH4+ and PO43– ions were estimated by colorimetric analysis using PhosVer HACH reagent and the salicylate method, respectively.
2.2 DNA extraction and AFLP procedure
The leaf material was homogenised in a TissueLyser LT bead mill (Qiagen) and used for DNA extraction with an AX Plant Kit (A&A Biotechnology) according to the manufacturer’s instructions. The AFLP procedure was as described by Vos et al. (1995) with some modifications suggested by Applied Biosystems (AFLP Plant Mapping Protocol). First, 56 primer combinations were tested on two Polish and two Finnish individuals to choose the four most polymorphic and repeatable primer pairs. The highest quality was shown by EcoRI-ACA/MseI-CTC, EcoRI-ACA/MseI-CTG, EcoRI-AGG/MseI-CAC and EcoRI-ACG/MseI-CAA combinations. The fluorescently (FAM or JOE) labelled amplification products were mixed with a GeneScan 500 LIZ Size Standard (Applied Biosystems) and run on an ABI PRISM 3130 (Applied Biosystems) instrument. AFLP genotypes were identified based on the presence (1) or absence (0) of bands between 70 and 500 bp using GENEMAPPER 4.0 software (Applied Biosystems). Only variable fragments were chosen. To minimise technique-related and scoring errors, the AFLP profiles were tested by replication of two individuals from each population starting from the restriction/ligation reaction. The error rate was estimated by the method recommended by Bonin et al. (2004).
2.3 Germination experiment
The total wet mass of 100 seeds per individual studied was measured with an accuracy of 0.0001 g (Table 2). To break dormancy, seeds were maintained at +4 °C from 1st December until mid-January and from 1st March until 31st March, and at –20 °C from mid-January until 1st March. After vernalisation, a germination test was conducted in a phytotron at a constant temperature of 20 °C and a photoperiod of 10 hrs light/14 hrs dark (Holm 1994). During this experiment seeds were placed in closed Petri dishes on wet filter paper. Petri dishes were examined every day to count germinated seeds (having visible radicles) and to remove them. Because the number of germinated seeds was extremely small (0.2%), we decided to examine the non-germinated seeds under a binocular microscope with ×8 magnification using dissecting needles. During examination, the seeds were classified as: empty (without ovule), filled, partially filled or damaged by insects (Table 2). In total, 9977 seeds were tested in this way. Filled seeds having coats destroyed by dissecting needles (mechanical scarification) were again placed in a phytotron, and 100% of them germinated.
2.4 Statistical analyses
For each population, the number of polymorphic loci (Ploc), Nei’s gene diversity (Hj) and the inbreeding coefficient (F) were calculated as measures of genetic variation. The inbreeding coefficient was determined using a Bayesian approach (mixed Gibbs-Metropolis algorithm) implemented in I4A computer program (Chybicki et al. 2011) with 60 000 sampling steps after 10 000 burn-in updates. Prior values of beta-distribution were α = β = 1.0. The F calculations were preceded by detection and removing loci under selection (‘outlier’ loci) because they could influence the level of inbreeding. The outliers were identified based on log(Bayes Factor) > 2 and a corresponding posterior probability > 0.99 in the hierarchical Bayesian method implemented in the BayeScan software version 2.1 (Foll and Gaggiotti 2008). Next, Ploc and Hj parameters were estimated at the 5% level using a Bayesian method with uniform prior distribution of allele frequencies (Zhivotovsky 1999) implemented in AFLP-SURV version 1.0 (Vekemans et al. 2002).
To resolve the problem of whether the intra-population genotypic variation of B. nana is either a consequence of recombination resulting from sexual reproduction or accumulation of somatic mutations in vegetative ramets, the cladistic approach of matrix incompatibility analysis was used (Mes 1998). Matrix incompatibility counts (MICs) were determined with the help of the JACTAX jack-knifing option in PICA version 4.0 (Wilkinson 2001). In this approach, incompatibility means that all four possible combinations at two AFLP binary loci, i.e., 0/0, 0/1, 1/0, and 1/1, are found in a population. Because the simplest explanation of this phenomenon is sexual reproduction, the level of genotypic incompatibility is equivalent to the extent of meiotic recombination: higher MIC values evidence more frequent recombination events. Genotypes generated by somatic mutations do not contribute to the MIC. The MIC was tested with 1000 permutations using the model of empirical frequencies. In each round, genotypes having the highest contribution to the MIC were removed from the total sample and the procedure was repeated until the MIC was zero (situation when only genotypes differentiated by somatic mutations remain). Because of the low number of individuals, genotypic incompatibilities were not tested in Belarusian and Russian populations.
The most likely number of independent genetic clusters was assessed using two software packages: STRUCTURE 2.3.4 (Pritchard et al. 2000) and SAMOVA 2.0 (Spatial Analysis of MOlecular Variance; Dupanloup et al. 2002). In STRUCTURE, 10 independent runs examining the number of populations (between K1 and K8) with 500 000 iterations and 50 000 burn-in periods were performed. The analyses were based on the admixture model with correlated allele frequencies. The optimal K values were inferred from the maximum value of ∆K (Evanno et al. 2005) as implemented in STRUCTURE HARVESTER v0.6.93 (Earl and vonHoldt 2012). SAMOVA allows the definition of groups of populations (K) that are geographically homogeneous and maximally differentiated from each other. Because contributions of one or more single-population groups mean that the group structure is disappearing (Magri et al. 2006), tests for K = 2 to K = 4 were performed for the eight populations of B. nana. Analysis of molecular variance (AMOVA) using ARLEQUIN version 3.11 (Excoffier et al. 2005) was then conducted to test the genetic relationships between and within the groups defined by SAMOVA. Genetic differentiation (FST) among pairs of populations was estimated with the help of ARLEQUIN. Genetic relationships between individuals were identified by principal component analysis (PCA) using the PAleontological STatistics (PAST) software version 3.14 (Hammer et al. 2001). The program PAST with 1000 bootstrap over 240 AFLP loci was also used to calculate Jaccard’s similarity coefficient (Hammer et al. 2001) between individuals and draw the resulting neighbour joining (NJ) tree. Only bootstrap values ≥ 50% were placed at the nodes of this tree.
Sexual reproduction performance of the Polish and Finnish populations of B. nana was evaluated based on the median values of the number of flowers as well as the mass, quality and germination capacity of seeds (Table 2). A two-sample randomisation test (10 000 randomisations) implemented in Rundom Pro 3.14 software (Jadwiszczak 2009) was used to compare the relict and central localities in terms of the total number of flowers per flowering individual, numbers of female and male flowers per flowering individual, wet seed mass, germination ability of seeds without (GS) and after scarification (GSC), and the contribution of empty (ES), partly filled (PF) and infected (IN) seeds. The Spearman’s correlation coefficient was calculated to infer a monotonic relationship between the seed mass and the number of germinated seeds (GS + GSC). A randomised one-way ANOVA (including post hoc tests and Holm’s correction; test statistic – mean difference; 10 000 randomisations) was conducted using Rundom Pro to compare chemical habitat factors (pH, EC, NH4+, PO43–) among the populations studied. Sequential Bonferroni correction (Rice 1989) was applied to the results of multiple tests.
3 Results
In total, 240 polymorphic AFLP loci were included in the genetic analyses (39 from EcoRI-ACA/MseI-CTC, 71 from EcoRI-ACA/MseI-CTG, 67 from EcoRI-AGG/MseI-CAC and 63 from EcoRI-ACG/MseI-CAA). The error rate was 3.4%. The highest contribution of polymorphic loci (Ploc) was recorded in the IZER population and the lowest in the LIN reserve, both situated in Poland (Table 1). The Nei’s gene diversity parameter (Hj) ranged from 0.091 in Tuusniemi (TUS) in Finland to 0.136 in the IZER. In all populations, mean inbreeding coefficient (F) values were higher than 0(Table 1). Both the minimum and maximum F values were noted in the Finnish populations TUS (0.035) and Kaitaa (KAI; 0.402), respectively. Inbreeding coefficients were calculated after removing one AFLP locus because it was likely under selection pressure [log(Bayes Factor) = 3.5227, P = 0.9997]. The MICs revealed that most genotypes in Polish and Finnish populations were formed by meiotic recombination. Participation of a single genotype in overall incompatibility was quite low; therefore, the successive deletion of the genotypes with the highest contributions generated a gradual loss of the MIC (Fig. 2).
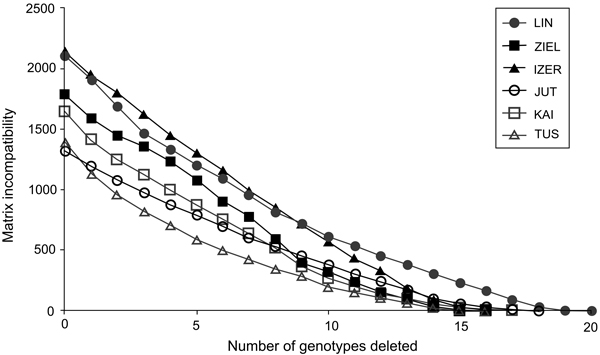
Fig. 2. The graph of matrix incompatibility counts (MICs). Reduction of MIC is related to the successive removal of genotypes from each of the specific population. Graphic symbols refer to codes of populations described in Table 1.
SAMOVA analyses revealed single-population sets for K = 3 (ZIEL and LIN were classified as two distinct sets, and all remaining populations were classified as a third group) and K = 4 (LIN, ZIEL and IZER were classified as three independent groups and all other populations were classified as a fourth group). The only combination including at least two populations in one group was for K = 2. In this combination, LIN and ZIEL localities formed one group and all remaining samples were included in a second group. AMOVA showed statistically significant genetic differentiation between LIN and ZIEL populations from Poland and all other samples (Table 3). Differentiation between populations within groups was highly significant (FSC = 0.18153, P < 0.0001); however, most of the genetic variation was found within populations (FST = 0.28684, P < 0.0001). All results of the pairwise comparisons of FST were significant after Bonferroni’s correction except for Berezen’skij Zapovednik (BZ) and Ural (UR) samples (Table 4). The K statistics of Evanno et al. (2005) revealed the maximum value for K = 2. In general, all Polish samples were assigned to one genetic cluster and all central populations and BZ from Belarus to another cluster (Fig. 3). However, the relict IZER population was assigned by STRUCTURE to the first cluster with P = 0.52 and to the second group with P = 0.48. In the PCA, grouping of the individual genotypes was similar to the pattern shown by STRUCTURE. The multilocus genotypes from the Polish populations were more diverse and most of them were separated from the remaining specimens, although mixing among two groups appeared to some extent (Fig. 4). Genotypes from the central populations Juttuvaara (JUT), KAI, TUS and UR as well as from the relict BZ were generally closely related one to another. In the PCA analysis the first axis explained 14.9% and the second 8.3% of the overall variance. Phylogenetic analysis using the NJ method also revealed genetic distinctness between the Polish and all remaining populations, although the bootstrap supports were below 50% (Fig. 5). In the biggest Polish cluster, some individuals from the KAI, BZ and UR samples were included. Almost all IZER specimens were nested into the two small groups, related more closely to the Polish samples than to the remaining ones.
| Table 3. Results from analyses of molecular variance in the B. nana populations divided into two groups: LIN + ZIEL (Poland) and all other samples. | |||
| Source of variation | Percentage of variation | P | Fixation indices |
| Among groups | 12.87 | 0.032 | FCT = 0.12866 |
| Among localities within groups | 15.82 | < 0.0001 | FSC = 0.18153 |
| Within localities | 71.32 | < 0.0001 | FST = 0.28684 |
| Table 4. Genetic differentiation (FST) between pairs of populations of B. nana. Population codes according to Table 1. * = statistically significant values after Bonfferoni’ s correction. | |||||||
| ZIEL | IZER | JUT | KAI | TUS | BZ | UR | |
| LIN | 0.244* | 0.271* | 0.314* | 0.196* | 0.339* | 0.228* | 0.278* |
| ZIEL | 0.234* | 0.359* | 0.249* | 0.367* | 0.285* | 0.301* | |
| IZER | 0.246* | 0.159* | 0.252* | 0.222* | 0.261* | ||
| JUT | 0.049* | 0.051* | 0.161* | 0.203* | |||
| KAI | 0.085* | 0.097* | 0.136* | ||||
| TUS | 0.176* | 0.217* | |||||
| BZ | 0.037 | ||||||
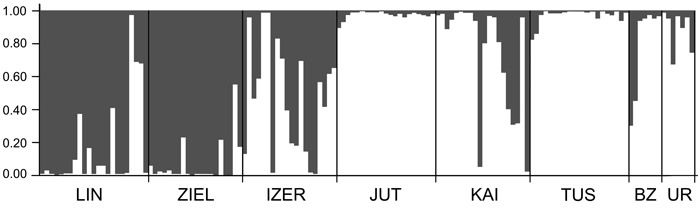
Fig. 3. Clustering results for the B. nana populations (K = 2) generated by STRUCTURE software. Population codes according to Table 1.
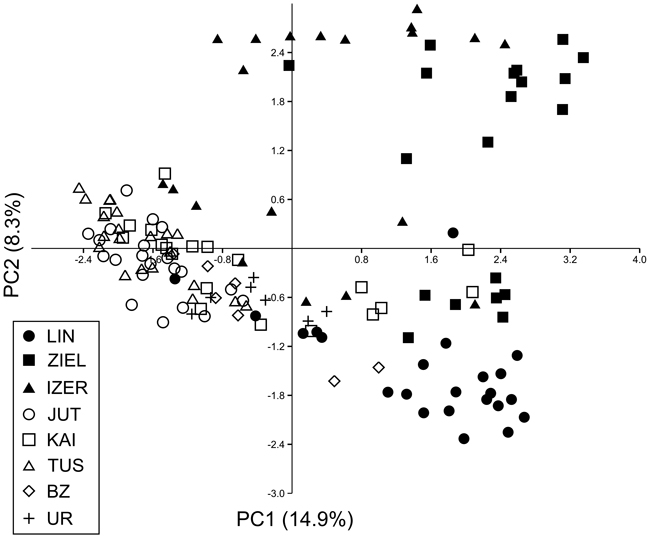
Fig. 4. Principal component analysis (PCA) plot revealing the genetic distances among 139 individuals of B. nana. Graphic symbols refer to codes of populations described in Table 1.
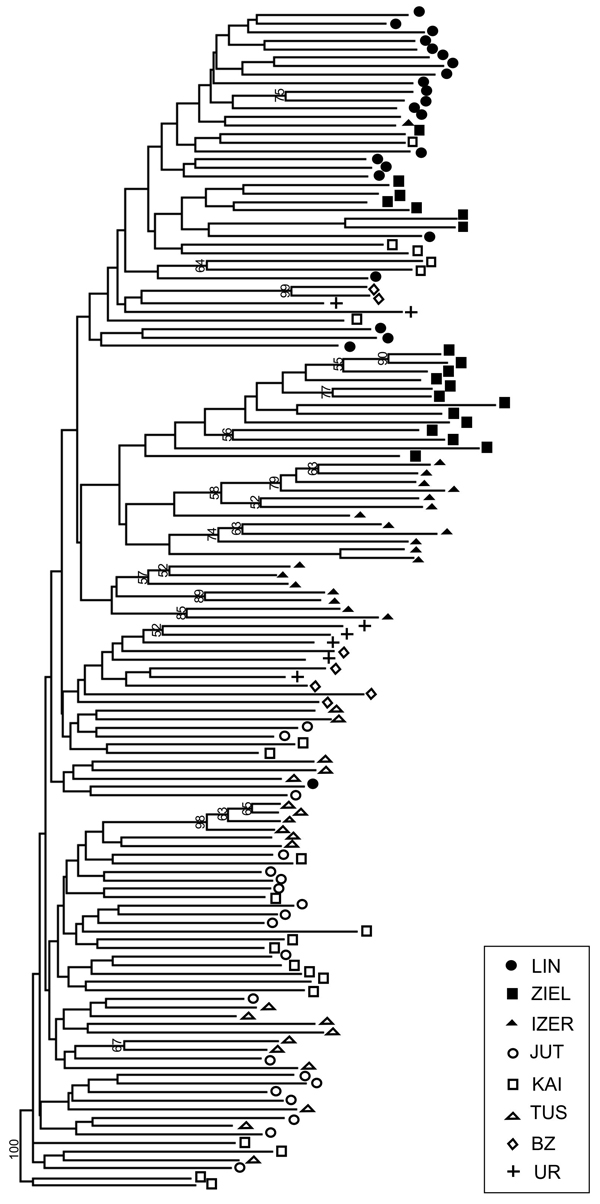
Fig. 5. Neighbour-joining tree based on 240 AFLP fragments comprising 8 populations of B. nana (based on Jaccard’s similarity coefficient; Hammer et al. 2001). Numbers on branches represent bootstrap support (1000 replicates). Graphic symbols refer to codes of populations described in Table 1.
All median values of the reproductive output parameters [the total number of flowers per flowering individual, numbers of female and male flowers per flowering individual, seed mass, germination ability of seeds without (GS) and after scarification (GSC), and number of empty (ES), partly filled (PF) and infected by insects (IN) seeds] are given in Table 2. After Bonferroni’s correction, three statistically significant results of the two-sample randomisation tests comparing the different parameters of sexual reproduction in the relict and central populations were found: the number of male flowers and seed mass were higher in the relict populations, and the number of seeds germinated after scarification was higher in central locations (Table 5). The correlation analysis did not demonstrate any relationship between the seed mass and total number of germinated seeds, e.g., without and after scarification (rs = 0.0955, P = 0.3447).
| Table 5. Results of the two-sample randomisation tests comparing the reproductive parameters (dependent variable) in the relict (R) vs. central (C) populations (grouping variables). * = value significant after Bonferroni’s correction. | ||||
| Variable | Test statistic | P | ||
| Grouping | Dependent | Medians | ||
| Relict vs. central samples | Female flowers | R = 1.0000 | 1.0000 | 0.3956 |
| C = 0.0000 | ||||
| Male flowers | R = 6.0000 | 3.0000 | 0.0074* | |
| C = 3.0000 | ||||
| Total no. of flowers | R = 7.5000 | 3.0000 | 0.0161 | |
| C = 4.5000 | ||||
| Seed mass [g] | R = 0.0159 | 0.0027 | 0.0035* | |
| C = 0.0132 | ||||
| Seeds germinated without scarification | R = 0.0000 | 0.0000 | 1.0000 | |
| C = 0.0000 | ||||
| Seeds germinated after scarification | R = 0.5000 | –3.5000 | 0.0002* | |
| C = 4.0000 | ||||
| Empty seeds | R = 95.5000 | 0.5000 | 0.7345 | |
| C = 95.0000 | ||||
| Partly filled seeds | R = 0.0000 | 0.0000 | 1.0000 | |
| C = 0.0000 | ||||
| Seeds infected by insects | R = 1.0000 | 0.0000 | 1.0000 | |
| C = 1.0000 | ||||
The means of chemical parameters (pH, EC, NH4+, PO43–) of habitat quality are shown in Table 2. One-way ANOVA analyses displayed that the two relict populations IZER and LIN had markedly different water characteristics (Table 6). IZER was significantly different from LIN, ZIEL, KAI and TUS for pH (F = 9.768, P = 0.0001); in turn, EC in the LIN sample differed strongly from all other populations (F = 9.293, P = 0.0001). Additionally, LIN and IZER were substantially different from some Polish and Finnish populations considering the concentrations of PO43– ions (F = 8.696, P = 0.0001).
| Table 6. Results of ANOVA comparing the chemical parameters of environment in the populations of B. nana. Populations codes according to Table 1. Padj = P adjusted. * = value significant after Bonferroni’s correction. | |||||
| Variable | DF | F | P | Post hoc tests | |
| pH | 52 | 9.768 | 0.0001* | IZER vs. TUS | Padj = 0.0015 |
| IZER vs. LIN | Padj = 0.0028 | ||||
| IZER vs. KAI | Padj = 0.0026 | ||||
| IZER vs. ZIEL | Padj = 0.0432 | ||||
| Electrical conductivity (EC) | 52 | 9.293 | 0.0001* | LIN vs. ZIEL | Padj = 0.0015 |
| LIN vs. JUT | Padj = 0.0070 | ||||
| LIN vs. KAI | Padj = 0.0065 | ||||
| LIN vs. TUS | Padj = 0.0072 | ||||
| LIN vs. IZER | Padj = 0.0297 | ||||
| NH4+ concentration | 52 | 1.924 | 0.1085 | - | - |
| PO43– concentration | 52 | 8.696 | 0.0001* | LIN vs. ZIEL | Padj = 0.0015 |
| LIN vs. TUS | Padj = 0.0014 | ||||
| ZIEL vs. IZER | Padj = 0.0065 | ||||
| IZER vs. TUS | Padj = 0.0084 | ||||
| LIN vs. JUT | Padj = 0.0209 | ||||
4 Discussion
Analysis of AFLP markers revealed that most genetic variation was found within populations of B. nana (AMOVA; FST = 0.28684, P < 0.0001), and relict stands from Poland and Belarus were not genetically depauperate compared with the central localities from Finland and Russia. The number of polymorphic loci was similar or higher (Ploc = 83–102) and Nei’s gene diversity parameter was higher (Hj = 0.120–0.136) in the relict samples than in samples derived from the widespread populations (Ploc = 87–91; Hj = 0.091–0.115). These results were most striking because nuclear microsatellite investigation has suggested low levels of intrapopulation genetic diversity in the Polish populations of B. nana (Jadwiszczak et al. 2012). It was explained as an effect of small population sizes, insufficient sexual reproduction and lack of gene exchange between the studied localities. However, the AFLP study showed that most genotypes in the B. nana populations arose as an effect of outcrossing. The MIC values were high and decreased gradually when individuals with the highest contribution to MIC were removed in succession. Only four or five genotypes in specific populations were formed by somatic mutations, which obviously indicated that meiotic recombination contributed to the overall genetic diversity of B. nana localities in Poland and Finland to a similar extent.
The next surprising outcome was the different rankings of Polish populations for genetic diversity estimated using the AFLPs and microsatellites. The highest and lowest levels of microsatellite variation were recorded in the LIN and IZER samples, respectively (Jadwiszczak et al. 2012). In turn, AFLPs were most variable in IZER and least variable in LIN. Diversity patterns obtained by microsatellite and AFLP systems were also poorly correlated at the population level in Pinus pinaster Aiton (Mariette et al. 2001), Eryngium alpinum L. (Gaudeul et al. 2004) and three Draba species (Skrede et al. 2009). Mariette et al. (2002) and Gaudeul et al. (2004) stated that lack of congruence between different molecular markers could result from uneven coverage of a genome by different marker systems, the differences in mutation rate of microsatellites and AFLPs, low heterogeneity of populations for their diversity and recently created populations. Considering the first explanation, it is obvious that in all studies the number of microsatellites used was much smaller than the AFLP loci. In the Polish populations of B. nana, 11 microsatellite loci (Jadwiszczak et al. 2012) and 240 AFLP loci were analysed; therefore, the coverage of the birch genome is considerably broader with AFLPs than with the microsatellites. More numerous markers appear to be more conclusive in the evaluation of population genetic diversity despite the codominant and multiallelic state of microsatellites (Mariette et al. 2002; Gaudeul et al. 2004). Additionally, the presence of null alleles, which can decrease the level of genetic variation assessed by the microsatellites, was not excluded at some microsatellite loci in the B. nana study (Jadwiszczak et al. 2012).
Discrepancy between AFLP and microsatellite results can also arise as an effect of the different mutation rates of both molecular systems. In general, the rate of mutation was found to be much faster for microsatellites (10–3–10–4 locus/generation) than for AFLP markers (10–6 locus/generation) (Mariette et al. 2001). However, it was observed in Pinus sylvestris L. inhabiting the Chernobyl exclusion zone that specific factors such as ionizing radiation could cause DNA damage across the entire genome and thus significantly increase the mutation rate at AFLP loci (Kuchma et al. 2011).
Simulations conducted by Mariette et al. (2002) indicated that low differentiation of populations for their diversity could be a reason for a lack of correlation between diversity parameters among markers. Low heterogeneity for diversity is observed in large-sized populations frequently exchanging genes. However, such explanation is not true for B. nana because the AMOVA analysis demonstrated significant genetic differentiation among populations within groups (FSC = 0.18153, P < 0.0001). We also rejected the hypothesis of Mariette et al. (2002) that the recent origin of samples could generate a discrepancy between values of genetic diversity parameters calculated from different markers. B. nana has occupied its stands in the Sudety Mts. since the Boreal period (9.0–7.5 ka) of the Holocene. The LIN locality is much older because the species has been there since the Younger Dryas (12.9–11.7 ka) of the Late Glacial (Jadwiszczak et al. 2012 and references therein).
Based on the nuclear microsatellite investigation, Polish populations of B. nana were recognised as locally critically endangered (Jadwiszczak et al. 2012). Although the AFLP analyses revealed that they represented a level of genetic variation comparable to that observed in the samples from the centre of the species range, we maintain that these relict locations should still be preserved. Genetic resources found in the relict stands can result from a long lifespan of genets, additionally enhanced by infrequent sexual reproduction, as was suggested for B. nana populations from the Svalbard archipelago (Alsos et al. 2002). Longevity of genets in clonal plants depends on successful clonal reproduction. B. nana is a polycormic shrub in which specific ramets are connected to the parent plant; however, they can exist as independent shoots when severed (Kummerow 1983). The oldest ramets in the LIN stands were 17 years old (Ejankowski 2008), although it has been found that sprouting in birches could occur from centuries-old root fragments (Brodie 1994). It is likely that clonal growth allows specific genets to survive in unsuitable habitat conditions and to produce flowers and seeds when conditions are relaxed. Hybridisation with close congeners can be a second possible explanation for the observation of comparable genetic variation in marginal and central B. nana localities. It was noticed that individuals in endangered populations, lacking from the mates of the same species, were more likely to hybridise with abundant congeners (Rhymer and Simberloff 1996). However, recent studies based on the nuclear microsatellites and AFLP fragments denied an extensive gene flow between B. nana and sympatric Betula pubescens Ehrh. (Wang et al. 2014; Eidesen et al. 2015).
Except for the UR and BZ samples, the values of genetic differentiation (FST) among populations of B. nana were statistically significant. Such results must be a consequence of the limitation or lack of gene flow among geographically isolated localities. Even Finnish populations were significantly different from each other, although they were physically most adjacent. A high degree of geographical isolation of the Polish populations prevents pollen and seed dispersal; these relict localities were thus considerably different from one another and from the other samples. Microsatellite investigation also indicated significant differentiation among Polish stands (Jadwiszczak et al. 2012). However, they were included in one genetic cluster by the STRUCTURE software in the present study. All remaining localities formed the second cluster. Note that incorporation of the IZER sample into the Polish group was not very explicit. First, the probability of the IZER membership in the Polish group was only slightly higher (P = 0.52) than the probability of its incorporation into the second cluster (P = 0.48). Second, the IZER individuals were clearly distinct from other Polish birches in the NJ tree. Third, some genotypes from the IZER sample were combined with the group formed by JUT, KAI, TUS, BZ and UR individuals in the PCA ordination. Fourth, SAMOVA analysis revealed a problem with group definition. A group identified by SAMOVA should consist of at least two populations that are geographically close and phylogeographically homogenous, whereas two groups should be maximally differentiated from each other (Shi et al. 2012). In our study, SAMOVA defined two groups, LIN and ZIEL from Poland, that were opposed to all other populations; the genetic differentiation among them was statistically significant (AMOVA; FCT = 0.12866, P = 0.032). The IZER sample was included in the second group, although it is located far from the closest BZ stand.
The weak parameters of sexual reproduction in the B. nana populations were rather prospective. It has been shown that the species reproduced mainly clonally in the Arctic localities (Alsos et al. 2003) and that sexual reproduction could be impaired in the LIN (Ejankowski 2004) as well as ZIEL and IZER (Jadwiszczak et al. 2012) stands, at least in some years. Clonal propagation is typical for Arctic and Alpine plants, which often grow in severe climate conditions with a deep litter layer that impedes the development of seedlings (Grime 2001; Ejankowski 2010).
Comparisons of reproductive parameters among relict and central stands of B. nana revealed that the number of male flowers and seed mass were significantly higher in Poland, whereas the number of GSC seeds was significantly higher in Finland. We think that the last result is more important in determining the sexual reproduction efficiency of the populations studied. First, the number of flowers (male and/or female) is not necessarily compatible with reproductive success. Flowering was abundant in the LIN locality in 2003, but inflorescences were later damaged by insects, which limited seed production (Ejankowski 2010). In the present study, we also noted insect activity, especially in LIN and TUS. Second, as the number of fully developed seeds was very low in both regions, seed mass was in fact the weight of seed coats. This was confirmed by the non-significant result of the Spearman’s correlation between the seed mass and the total number of GS and GSC seeds (rs = 0.0955, P = 0.3447). The lowest median value of empty seeds was recorded in TUS (77.5) and the highest values were noted in ZIEL, IZER, JUT and KAI, where almost all seeds were empty. Empty seeds are frequently found in birches. Holm (1994) discovered that 36–74% and 43–81% of seeds in northern Swedish populations of Betula pendula Roth and B. pubescens, respectively, were without ovules. X-ray analysis conducted in 10 relict populations of B. nana in Germany revealed no more than 13% of viable seeds per sample (Schwarz and Poschlod 2015). For empty seed production in angiosperm plants, either self-incompatibility or inbreeding can be responsible (Wang 2003). In our opinion, these factors are not very important in the B. nana locations because all samples were genotypically diverse. The inbreeding coefficients were increased in the KAI, LIN and ZIEL samples. At the same time one of the lowest numbers of empty seeds was recorded in the LIN population. It is likely that huge numbers of empty seeds can result from lack of pollination because such a phenomenon has been shown in birches (Atkinson 1992).
Based on the GSC parameter, we assume that the effectiveness of sexual reproduction of B. nana can be higher in the central populations compared with the relict stands. Greater seed germination ability has also been indicated as a determinant of better sexual reproduction of endangered Betula humilis Schrk. in the subcentral localities than in the marginal ones (Chrzanowska et al. 2016). However, it is difficult to say why some seeds were not able to germinate without mechanical destruction of their coats. Because frost treatment enhances germination ability in Arctic species (Alsos et al. 2013), one possible explanation is the too short time of frost treatment. B. nana seeds were maintained for one and a half months at –20 °C, which was much longer than in the more successful germination trial of B. humilis (Chrzanowska et al. 2016). On the other hand, seeds of B. nana var. tundrarum from the Arctic Archipelago of Svalbard were stored for almost five months at –14 °C and no seed germinated (Alsos et al. 2013). Another explanation is the presence of inhibitors in coats. It has been demonstrated that the intact seeds of B. pendula and B. pubescens require higher oxygen concentrations to sprout, whereas damaged coats probably enhance oxygen entry and quicker development of embryos (Black and Wareing 1959). It has also been revealed that germination was sometimes blocked by an impermeable seed coat (physical dormancy), without additional physiological reasons, as in the cases of the two Alpine species Anthyllis alpicola and Trifolium pallescens (Schwienbacher et al. 2011).
The efficiency of sexual reproduction of plants can change in relation to habitat conditions. The germination capacity of B. humilis seeds was significantly reduced at dry or more shaded sites compared to wet or sun-exposed sites (Chrzanowska et al. 2016). In the LIN population, shading by Ledum palustre and Vaccinium uliginosum and high water level were responsible for the low number of flowering ramets (Ejankowski 2008; 2010). Comparing plant communities supplied with phosphorus and nitrogen with those with ambient soil nutrients, Gough et al. (2015) found that B. nana started to produce more inflorescences after 5 years of N + P fertilisation and more seeds after 22 years of treatment. Increased soil nitrogen and organic matter contents were also proven to increase investment in flower production of circumpolar Bistorta vivipara (Bills et al. 2015). Analyses of chemical parameters of water in the B. nana locations suggested that the IZER and LIN samples were significantly different from other populations because of pH and EC measures, respectively. Both populations also showed considerably higher concentrations of PO43– ions compared to some other sites. According to Fujita et al. (2014), plants growing in N-limited habitats begin to flower earlier, have longer flowering time, more seeds and heavier seeds compared to plants inhabiting P-limited areas. Because sexual reproduction requires a substantial amount of P (Obeso 2002), it is likely that the highest number of male flowers per flowering individual noted at the LIN locality, and in consequence, the total number of flowers, was related to the higher PO43– concentrations at this site. Unfortunately, at the same time many seeds (median value of 7.5) in LIN were destroyed by insects.
Acknowledgements
We thank Piotr Jadwiszczak, Oleg Sozinov and Svetlana Pavlova who kindly helped to collect samples. Sample collection in Poland was approved by the Regional Directors of Environmental Protection in Wrocław (WPN.6205.59.2011.MR) and Bydgoszcz (WPN.6402.1.16.2011.JC). This study was funded by the National Science Centre, Poland (grant No. 2011/01/B/NZ8/01756).
References
Alsos I.G., Engelskjøn T., Brochmann C. (2002). Conservation genetics and population history of Betula nana, Vaccinium uliginosum, and Campanula rotundifolia in the Arctic archipelago of Svalbard. Arctic, Antarctic, and Alpine Research 34(4): 408–418. https://doi.org/10.2307/1552198.
Alsos I.G., Spjelkavik S., Engelsjøn T. (2003). Seed bank size and composition of Betula nana, Vaccinium uliginosum, and Campanula rotundifolia habitats in Svalbard and northern Norway. Canadian Journal of Botany 81(3): 220–231. https://doi.org/10.1139/b03-018.
Alsos I.G., Müller E., Eidesen P. (2013). Germinating seeds or bulbils in 87 of 113 tested Arctic species indicate potential for ex situ seed bank storage. Polar Biology 36(6): 819–830. https://doi.org/10.1007/s00300-013-1307-7.
Atkinson M.D. (1992). Betula pendula Roth (B. verrucosa Ehrh.) and B. pubescens Ehrh. Journal of Ecology 80(4): 837–870. https://doi.org/10.2307/2260870.
Bills J.W., Roalson E.H., Busch J.W., Eidesen P.B. (2015). Environmental and genetic correlates of allocation to sexual reproduction in the circumpolar plant Bistorta vivipara. American Journal of Botany 102(7): 1174–1186. https://doi.org/10.3732/ajb.1400431.
Black M., Wareing P.F. (1959). The role of germination inhibitors and oxygen in the dormancy of the light-sensitive seed of Betula species. Journal of Experimental Botany 10(1): 134–145. https://doi.org/10.1093/jxb/10.1.134.
Bolnick D.I., Amarasekare P., Araujo M.S., Burger R., Levine J.M., Novak M., Rudolf V.H.W., Schreiber S.J., Urban M.C., Vasseur D.A. (2011). Why intraspecific trait variation matters in community ecology. Trends in Ecology and Evolution 26(4): 183–192. https://doi.org/10.1016/j.tree.2011.01.009.
Bonin A., Bellemain E., Eidesen P.B., Pompanon F., Brochmann C., Taberlet P. (2004). How to track and assess genotyping errors in population genetics studies. Molecular Ecology 13(11): 3261–3273. https://doi.org/10.1111/j.1365-294X.2004.02346.x.
Brodie I. (1994). Management guidelines for birch. The Birch Improvement Company, Cuilalunn, Scotland.
Chrzanowska A., Jadwiszczak K.A., Kłosowski S., Banaszek A., Sozinov O.V. (2016). Sexual reproduction efficiency and genetic diversity of endangered Betula humilis Schrk. populations from edge and sub-central parts of its range. Folia Geobotanica 51(2): 161–173. https://doi.org/10.1007/s12224-016-9244-1.
Chybicki I.J., Oleksa A., Burczyk J. (2011). Increased inbreeding and strong kinship structure in Taxus baccata estimated from both AFLP and SSR data. Heredity 107: 589–600. https://doi.org/10.1038/hdy.2011.51.
Dupanloup I., Schneider S., Excoffier L. (2002). A simulated annealing approach to define the genetic structure of populations. Molecular Ecology 11(12): 2571–2581. https://doi.org/10.1046/j.1365-294X.2002.01650.x.
Earl D.A., vonHoldt B.M. (2012). Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4(2): 359–361. https://doi.org/10.1007/s12686-011-9548-7.
Eidesen P.B., Alsos I., Brochmann C. (2015). Comparative analyses of plastid and AFLP data suggest different colonization history and asymmetric hybridisation between Betula pubescens and B. nana. Molecular Ecology 24(15): 3993–4009. https://doi.org/10.1111/mec.13289.
Ejankowski W. (2004). The influence of ground water level on the demography and population structure of the dwarf birch Betula nana L. Ecological Questions 6: 63–68.
Ejankowski W. (2008). Effect of waterlogging on regeneration in the dwarf birch (Betula nana). Biologia 63(5): 670–676. https://doi.org/10.2478/s11756-008-0126-8.
Ejankowski W. (2010). Demographic variation of dwarf birch (Betula nana) in communities dominated by Ledum palustre and Vaccinium uliginosum. Biologia 65(2): 248–253. https://doi.org/10.2478/s11756-010-0007-9.
Ellstrand N.C., Elam D.R. (1993). Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology, Evolution, and Systematics 24: 217–242. https://doi.org/10.1146/annurev.es.24.110193.001245.
Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14(8): 2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x.
Excoffier L., Laval G., Schneider S. (2005). Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47–50.
Foll M., Gaggiotti O. (2008). A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180(2): 977–993. https://doi.org/10.1534/genetics.108.092221.
Fujita Y., Venterink H.O., van Bodegom P.M., Douma J.C., Heil G.W., Hölzel N., Jabłońska E., Kotowski W., Okruszko T., Pawlikowski P., de Ruiter P.C., Wassen M.J. (2014). Low investment in sexual reproduction threatens plants adapted to phosphorus limitation. Nature 505: 82–86. https://doi.org/10.1038/nature12733.
Gaudeul M., Till-Bottraud I., Barjon F., Manel S. (2004). Genetic diversity and differentiation in Eryngium alpinum L. (Apiaceae): comparison of AFLP and microsatellite markers. Heredity 92: 508–518. https://doi.org/10.1038/sj.hdy.6800443.
Gough L., Bass H., McLaren J.R. (2015). Effects of increased soil nutrients on seed rain: a role for seed dispersal in the greening of the Arctic? Arctic, Antarctic, and Alpine Research 47(1): 27–34. https://doi.org/10.1657/AAAR0014-055.
Grime J.P. (2001). Plant strategies, vegetation processes, and ecosystem properties. John Wiley & Sons, Chichester. 456 p. ISBN 978-0-470-85040-4.
Hammer Ø., Harper D.A.T., Ryan P.D. (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4(1) article 4. 9 p.
Hampe A., Petit R. (2005). Conserving biodiversity under climate change: the rear edge matters. Ecology Letters 8(5): 461–467. https://doi.org/10.1111/j.1461-0248.2005.00739.x.
Hampe A., Jump A.S. (2011). Climate relicts: past, present, future. Annual Review of Ecology, Evolution, and Systematics 42: 313–333. https://doi.org/10.1146/annurev-ecolsys-102710-145015.
Hewitt G.M. (2004). Genetic consequences of climatic oscillations in the Quaternary. Philosophical Transactions of the Royal Society B 359(1442): 183–195. https://doi.org/10.1098/rstb.2003.1388.
Holm S.-O. (1994). Reproductive patterns of Betula pendula and B. pubescens coll. along a regional altitudinal gradient in northern Sweden. Ecography 17(1): 60–72. https://doi.org/10.1111/j.1600-0587.1994.tb00077.x.
Hughes A.R., Inouye B.D., Johnson M.T.J., Underwood N., Vellend M. (2008). Ecological consequences of genetic diversity. Ecology Letters 11(6): 609–623. https://doi.org/10.1111/j.1461-0248.2008.01179.x.
Jadwiszczak K.A., Drzymulska D., Banaszek A., Jadwiszczak P. (2012). Population history, genetic variation and conservation status of the endangered birch species Betula nana L. in Poland. Silva Fennica 46(4) article 905. https://doi.org/10.14214/sf.905.
Jadwiszczak P. (2009). Rundom Pro 3.14. Software for classical and computer-intensive statistics. http://pjadw.tripod.com.
Jolly D., Harrison S.P., Damnati B., Bonnefille R. (1998). Simulated climate and biomes of Africa during the Late Quaternary: comparison with pollen and lake status data. Late Quaternary climates: data syntheses and model experiments. Quaternary Science Reviews 17(6–7): 629–657. https://doi.org/10.1016/S0277-3791(98)00015-8.
Kruszelnicki J., Herbichowa M., Kuczyńska M., Fabiszewski J. (2014). Betula nana L. Brzoza karłowata. [Betula nana L. Dwarf birch]. In: Kaźmierczakowa R., Zarzycki K., Mirek Z. (eds.). Polska czerwona księga roślin. [Polish plant red book]. W. Szafer Institute of Botany, Polish Academy of Sciences, Kraków. p. 95–97. ISBN 978-83-61191-72-8.
Kuchma O., Vornam B., Finkeldey R. (2011). Mutation rates in Scots pine (Pinus sylvestris L.) from the Chernobyl exclusion zone evaluated with amplified fragment-length polymorphisms (AFLPs) and microsatellite markers. Mutation Research 725(1–2): 29–35. https://doi.org/10.1016/j.mrgentox.2011.07.003.
Kummerow J. (1983). Root surface/leaf area ratios in Arctic dwarf shrubs. Plant Soil 71(1): 359–399. https://doi.org/10.1007/BF02182681.
Magri D., Vendramin G.G., Comps B., Dupanloup I., Geburek T., Gömöry D., Latałowa M., Litt T., Paule L., Rouge J.M., Tantau I., van der Knaap W.O., Petit R.J., de Beaulieu J.L. (2006). A new scenario for the quaternary history of European beech populations: palaeobotanical evidence and genetic consequences. New Phytologist 171(1): 199–221. https://doi.org/10.1111/j.1469-8137.2006.01740.x.
Mariette S., Chagné D., Lézier C., Pastuszka P., Raffin A., Plomion C., Kremer A. (2001). Genetic diversity within and among Pinus pinaster populations: comparison between AFLP and microsatellite markers. Heredity 86: 469–479. https://doi.org/10.1046/j.1365-2540.2001.00852.x.
Mariette S., Le Corre V., Austerlitz F., Kremer A. (2002). Sampling within the genome for measuring within-population diversity: trade-offs between markers. Molecular Ecology 11(7): 1145–1156. https://doi.org/10.1046/j.1365-294X.2002.01519.x.
Mes T.H.M. (1998). Character compatibility of molecular markers to distinguish asexual and sexual reproduction. Molecular Ecology 7(12): 1719–1727. https://doi.org/10.1046/j.1365-294x.1998.00508.x.
Obeso J.R. (2002). The costs of reproduction in plants. New Phytologist 155(3): 321–348. https://doi.org/10.1046/j.1469-8137.2002.00477.x.
Paun O., Schönswetter P., Winkler M., IntraBioDiv consortium, Tribsch A. (2008). Historical divergence vs. contemporary gene flow: evolutionary history of the calcicole Ranunculus alpestris group (Ranunculaceae) in the European Alps and the Carpathians. Molecular Ecology 17(19): 4263–4275. https://doi.org/10.1111/j.1365-294X.2008.03908.x.
Pritchard J.K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Provan J., Bennett K.D. (2008). Phylogeographic insights into cryptic glacial refugia. Trends in Ecology and Evolution 23(10): 564–571. https://doi.org/10.1016/j.tree.2008.06.010.
Pruett C.L., Winker K. (2008). Evidence for cryptic northern refugia among high- and temperate-latitude species in Beringia. A response to Stewart and Dalén (2008). Climatic Change 86(1): 23–27. https://doi.org/10.1007/s10584-007-9332-6.
Rhymer J.M., Simberloff D. (1996). Extinction by hybridization and introgression. Annual Review of Ecology and Systematics 27: 83–109. https://doi.org/10.1146/annurev.ecolsys.27.1.83.
Rice W.R. (1989). Analyzing tables of statistical tests. Evolution 43(1): 223–225. https://doi.org/10.2307/2409177.
Schwartz B.U., Poschlod P. (2015). Die Letzten ihrer Art in Bayern – Das Eiszeitrelikt Zwergbirke (Betula nana L.). Eine Bestandsanalyse mit biologisch-ökologischen Untersuchungen. [The last of its kind in Bavaria – the ice age relict Betula nana. Results of biological, ecological, and population viability studies]. Anliegen Natur 37. 12 p.
Schwienbacher E., Navarro-Cano J.A., Neuner G., Erschbamer B. (2011). Seed dormancy in alpine species. Flora 206(10): 845–856. https://doi.org/10.1016/j.flora.2011.05.001.
Shi W., Kerdelhue C., Ye H. (2012). Genetic structure and inferences on potential source areas for Bactrocera dorsalis (Hendel) based on mitochondrial and microsatellite markers. PLoS One 7(5): e37083. https://doi.org/10.1371/journal.pone.0037083.
Skrede I., Borgen L., Brochmann C. (2009). Genetic structuring in three closely related circumpolar plant species: AFLP versus microsatellite markers and high-arctic versus arctic–alpine distributions. Heredity 102: 293–302. https://doi.org/10.1038/hdy.2008.120.
Stewart J.R., Lister A.M. (2001). Cryptic northern refugia and the origins of modern biota. Trends in Ecology and Evolution 16(11): 608–613. https://doi.org/10.1016/S0169-5347(01)02338-2.
Vekemans X. (2002). AFLP-SURV. Version 1.0. Distributed by the author, Laboratoire Génétique et Ecologie Végétale, Université Libre de Bruxelles, Belgium.
Vos P., Hoger R., Bleeker M., Reijans M., van de Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M., Zabeau M. (1995). AFLP: a new technique for DNA ingerprinting. Nucleic Acids Research 23(21): 4407–4414. https://doi.org/10.1093/nar/23.21.4407.
Wang K.S. (2003). Relationship between empty seed and genetic factors in European beech (Fagus sylvatica L.). Silva Fennica 37(4) article 481. https://doi.org/10.14214/sf.481.
Wang N., Borrell J.S., Bodles W.J.A., Kuttapitiya A., Nichols R.A., Buggs R.J.A. (2014). Molecular footprints of the Holocene retreat of dwarf birch in Britain. Molecular Ecology 23(11): 2771–2782. https://doi.org/10.1111/mec.12768.
Wilkinson M. (2001). PICA 4.0: software and documentation. Department of Zoology, The Natural History Museum, London.
Zhivotovsky L.A. (1999). Estimating population structure in diploids with multilocus dominant DNA markers. Molecular Ecology 8(6): 907–913. https://doi.org/10.1046/j.1365-294x.1999.00620.x.
Total of 59 references.

