Spatial genetic structure and clonal structure of Prunus serotina during invasive spread
Dering M., Sękiewicz K., Iszkuło G., Chojnacka A., Tomaszewski D., Pers-Kamczyc E., Karolewski P. (2018). Spatial genetic structure and clonal structure of Prunus serotina during invasive spread. Silva Fennica vol. 52 no. 3 article id 9987. https://doi.org/10.14214/sf.9987
Highlights
- The spread of Prunus serotina in invaded forests is facilitated by high propagule pressure
- The seed shadow overlap prevents strong spatial genetic structure
- During colonization of isolated site, a strong spatial genetic structure is produced due to founder effect
- Overall clonality in P. serotina was low but may efficiently support seedling bank thus contributing to species invasiveness.
Abstract
Prunus serotina Ehrh. (black cherry) is one of the most important invaders in the European forests, but existing studies have given limited insight into demo-genetic factors underpinning the process of species invasion. Fine-scale genetic structure (FSGS) may deliver important knowledge on genetics of invasion contributing to efficient management of the alien species. Using eight microsatellites we investigated FSGS, clonal structure and relatedness in four black cherry populations which represented different stages of the invasive spread into Scots pine forests. Three populations were in a continuous forest complex and represented the colonization (Z_1) and established stages (Z_2 and Z_3). To investigate how colonization ability of the species is modified by landscape features, we analyzed an isolated population at colonization stage located in limited forest patch located in an agricultural landscape (R). Populations from continuous forest showed low yet significant positive FSGS with Sp = 0.0068 in Z_1, 0.0054 in Z_2, and 0.0066 in Z_3, while in R spatial structure was the strongest (0.0145). Considerable relatedness noted in population R suggests a dominance of within-population mating and recruitments, low immigration rate and limited seed dispersal, all of which led to the observed strong FSGS. Also, we presume that a founder effect likely involved during colonization of isolated forest patch R led to strong FSGS. In contrary, the seed shadow overlap in the populations from continuous forest prevented strong FSGS and facilitated colonization. Despite of low level of clonality, we argue that it may efficiently support black cherry seedling bank contributing to species invasiveness.
Keywords
genetic structure;
invasiveness;
black cherry;
colonization;
clonal structure
-
Dering,
Institute of Dendrology, Polish Academy of Sciences, Parkowa 5, 62-035 Kórnik, Poland
E-mail
mdering@man.poznan.pl

- Sękiewicz, Institute of Dendrology, Polish Academy of Sciences, Parkowa 5, 62-035 Kórnik, Poland E-mail ksekiewicz@man.poznan.pl
- Iszkuło, Institute of Dendrology, Polish Academy of Sciences, Parkowa 5, 62-035 Kórnik, Poland; Faculty of Biological Sciences, University of Zielona Góra, Prof. Z. Szafrana 1, 65-516 Zielona Góra, Poland E-mail iszkulo@man.poznan.pl
- Chojnacka, Institute of Dendrology, Polish Academy of Sciences, Parkowa 5, 62-035 Kórnik, Poland E-mail jagoda900@gmail.com
- Tomaszewski, Institute of Dendrology, Polish Academy of Sciences, Parkowa 5, 62-035 Kórnik, Poland E-mail dominito@man.poznan.pl
- Pers-Kamczyc, Institute of Dendrology, Polish Academy of Sciences, Parkowa 5, 62-035 Kórnik, Poland E-mail epk@man.poznan.pl
- Karolewski, Institute of Dendrology, Polish Academy of Sciences, Parkowa 5, 62-035 Kórnik, Poland E-mail pkarolew@man.poznan.pl
Received 30 March 2018 Accepted 27 July 2018 Published 13 August 2018
Views 120041
Available at https://doi.org/10.14214/sf.9987 | Download PDF

1 Introduction
Human-mediated introductions of plants and animals have a very long history, but only since the 20th century with increased globalization has the redistribution of species accelerated and become a global issue both in scale and consequences. Biological invasions cause global homogenization of biodiversity, which challenges the biogeographic patterns we know (Capinha et al. 2015), affect ecosystem processes and services that support human societies (Aerts et al. 2017; Craven et al. 2017; Taylor et al. 2017), may generate great costs for local and national budgets and even stir up social conflicts (van Wilgen et al. 2008; Crowley et al. 2017). In the context of ongoing climate changes, biological invasions emerge as a serious problem that the global ecosystem will have to deal with (Hulme 2017).
Forestry is a human activity that has actively contributed to the mass introduction of alien tree species all over the world in order to increase timber yields (Águas et al. 2017). Some of these tree species have spread from plantations into adjacent habitats, converted into successful invaders. A striking example is the use of pines on an unprecedented scale in commercial plantations in the Southern Hemisphere which has become a serious issue due to the wide-ranging negative influences they exert on the functioning and structure of the invaded ecosystems (Bravo-Monasterio et al. 2016; Mostert et al. 2017). Quercus rubra L. was introduced from North America into European forestry in the early 19th century and has recently been recognized as an invasive species threatening native biodiversity (Riepšas and Straigytė 2008; Woziwoda et al. 2014; Tyler et al. 2015). But spectacular invasions do not necessarily involve transcontinental transfers. Pinus pinaster Aiton, native to the Mediterranean, has been widely used in afforestation beyond its natural range across the Mediterranean countries, but has turned out to be a very aggressive invader, able to displace natural evergreen and deciduous woodlands leading to a decrease in taxonomic and phylogenetic diversity of the invaded areas (Selvi et al. 2016).
The process of plant invasion can be framed in a model consisting of a few consecutive stages such as transport from the natural range, introduction into a new territory, establishment, and finally invasive spread into other sites within the new range (Blackburn et al. 2011). The three first stages (transport, introduction and establishment) are of interest to many researchers aiming to explore the exact sources and subsequent pathways of species invasions. However, recognition of the mechanisms and factors present during invasive spread at new sites matters greatly for the identification of population genetic processes underlying invasion’s successes and failures. To get the full picture of an invasion process it also seems particularly necessary to study the process across invasion stages (e.g. colonizing and established population), environmental and landscape complexity or habitat features (Berens et al. 2014). A deep understanding of invasive genetics in a wider ecological perspective may be used for tailoring efficient management strategies for alien invasive species.
Invasion biology attempts to understand the process of invasion by asking about the causes and conditions of the successful invasions. From one side, the attention is paid on the external factors linked to the ecological and human-related aspects of the invasions, which has been enclosed in a term of a community invasibility (Richardson and Pyšek 2006); from the other side, studies also try to profile a successful invader by pointing the specific set of traits that is shared among the most successful invaders (Pyšek and Richardson 2008). Studies confirmed that the traits do really matter, and those related with the propagules – or generally with reproduction – matter greatly. Invasive ecology studies repeatedly underline that clonality is positively associated with species invasiveness (Sakai et al. 2001; Liu et al. 2006; Kelager et al. 2013; Gaskin et al. 2014; Thomas et al. 2016). This life-history trait allows survival in the very first stages of the invasion when immigrants face with the new environments (Okada et al. 2009) or gives the competitive superiority to congeneric native species, which also facilitate invasion (Travis et al. 2011). In their theoretical model, Rafajlović et al. (2017) indicated that dioecious clonal species may form a “asexual colonization wave” during colonization that efficiently supports the establishment of recent invaders, especially in situation of the biased sex ratio. In extreme conditions, clonality even enables wide colonization by one sex only (Budde et al. 2011), and in the common garden experiment, Castro et al. (2016) reported a transition into clonality in reproductive strategy in plants from the invasive range. All these studies clearly show that clonality is a very important trait in plant invaders.
Prunus serotina Ehrh. (black cherry) is considered one of the “100 of the Worst” invasive species (DAISIE; www.europe-aliens.org). It was introduced to Europe from the north-eastern United States in the 17th century as an ornamental tree and was used in the 19th century in forestry for timber production, with low levels of success. Because it was expected to be useful for soil improvement, and wind- and firebreaks, black cherry was intensively grown in European forests during the first half of the 20th century (Lorenz et al. 2004). Currently, the species is naturalized but mostly invasive in much of Central Europe where it typically invades Scots pine-dominated stands. Recent studies have clearly showed the adverse effects of black cherry invasions in forest stands including limiting recruitment of native species (Vanhellemont et al. 2010), shifting forest community structure (Halarewicz and Żołnierz 2014) or even changing ecosystem functioning by altering nutrient cycling leading to a reduced carbon sequestration capacity of native species (Aerts et al. 2017). Leaves of P. serotina are successfully used as an alternative food source for native insect pests (Karolewski et al. 2014) that increases the food base for the insect pests of the native bird cherry (Prunus padus L.). Currently, there is no efficient method for invader eradication.
Pairon et al. (2010) reconstructed the introduction history of black cherry to Europe and indicated that there were several introduction events, which prevented the species from reduction of genetic diversity (Petitpierre et al. 2009); a significant bottleneck was detected only in the populations at the limits of the expanding range (Pairon et al. 2010). These results appreciably contribute to our understanding of the initial stages of the invasion process (transport, introduction, establishment of a new range) in this species, but we need a knowledge on the mechanisms acting during the invasive spread to successfully deal with the invasion. Since clonality is one of the important trait of the invasiveness syndrome, a reliable data on P. serotina ability of clonal reproduction and its impact on the species genetic structure and dynamics is highly required. Is clonality a significant factor during colonization of a new territories by this species? What is the clonal structure of the invaded forests and how it changes during invasive spread, if it changes at all? This has not been yet thoroughly studied and the existing data concerning overall clonality in P. serotina are very limited (Pairon et al. 2006; Jagodziński et al. 2015).
Studies exploring changes in fine-scale spatial genetic structure (FSGS) during invasive spread are rare (Sloop et al. 2011; Barnaud et al. 2013), and none has been conducted for P. serotina. Meanwhile, results of FSGS are commonly used in reconstructions of demographical and colonization histories of plant populations (Jones et al. 2006; Chybicki et al. 2011; Dering et al. 2016) and can provide information on the origin and number of invasion founders and events, deliver details on the demographic history and population genetic structure of invaders, and unravel the evolutionary genetics of invasions with special regard to the fast adaptability of invaders (Pairon et al. 2010; Blackburn et al. 2015; Bock et al. 2015). By setting daughter ramets in a space, clonal reproduction affects the disutibution of genotypes in a space, which affects the FSGS in a population. Study which compared clonal tree species with non-clonal species showed that spatial genetic structure estimated at the genet level (i.e., without repeted genotypes) was almost three times higher in clonal trees than in non-clonal ones (Dering et al. 2015). The inclusion of ramets into estmation of spatial structure generally leads to its intensification (Dering et al. 2016). For example, estimation of FSGS in wild cherry (Prunus avium L.) resulted in high values of FSGS at the genet level (Sp ranging from 0.030 to 0.045), but FSGS at the ramet level was even twice as higher (Sp ranging from 0.089 to 0.119). Strong FSGS may have profound evolutionary consequences for plant popualtions because it may lead to biparental inbreeding, which results in a decrease of genetic diversity. Hence, FSGS may serve as the valuable source on the microevolutionary processes acting at different stages of the process of the biological invasion (Barnaud et al. 2013).
In this study, we aimed to investigate the population genetic patterns underlying the invasive spread of P. serotina in Pinus sylvestris L. stands. Hence, we compared FSGS between (1) populations at colonization and established stage and (2) between populations at colonization stage located in continuous forest complex and in agricultural environment. Given the results suggesting limited seed dispersal in black cherry (Pairon et al. 2006), we expected (1) to find stronger FSGS and higher overall relatedness in the populations at the colonization stage than in established populations and (2) to observe higher FSGS in population at colonization stage located in the agricultural landscape in comparison to population from the extensive forest complex due to spatial isolation and stronger founder effect. Black cherry is capable of clonal growth by root suckering (Auclair 1975), but existing studies have given limited insight into the role of this trait in species population dynamics, genetic structure and invasiveness (Closset-Kopp et al. 2007). Hence, in this study we also aimed to assess the level of clonality, assuming that (1) clonal recruitment dominates in population dynamics and thus this trait contributes significantly to species invasiveness and (2) clonality intensifies FSGS in black cherry populations.
2 Material and methods
2.1 Study sites and sampling design
We studied populations of black cherry located in P. sylvestris stands in the Zielonka Forest complex (Poland). In this forest complex, covering the total area of over 12 000 ha, P. serotina was introduced from 1949 up to the 1960s as an understory species and has been spreading ever since. The investigated populations of P. serotina represent spontaneous invasion into Scots pine stands of artificial origin. In contrary to short-lived species with high rates of population turnover, direct tracing of the invasion process starting from the colonization stage up to fully established population of an invader in case of tree species remains as genuine problem due to longevity of trees. To overcome such constrains, we adopted an indirect method. During field inspection, we chose three populations of black cherry, Z_1, Z_2 and Z_3, in which the age of the 10 individuals with the biggest trunk circumference was assessed dendrochronologically. Wood samples (Z_1 discs, Z_2 and Z_3 cores using increment borer) were taken from the bottom of the trunk. The samples were scanned and age analysis was conducted using a WinDENDRO Image Analyzing System (Regent Instruments Inc.). Homogeneity of ring series was subsequently verified using the COFECHA software (Holmes 1994). Based on the obtained average age, populations were assigned to one of the two successional stages: Z_1 – colonization and Z_2 and Z_3 – established (Table 1). Stands Z_1 and Z_2 were pure, one forest-floor Pinus sylvestris stands, while population Z_3 had a second forest-floor with Quercus robur L. at age of 80 years (ca. 7% of the stand). Additionally, to investigate invasive colonization in different landscape context, a single population of black cherry located in an isolated Pinus sylvestris stand in Raszków was included (R). The age of the oldest individuals was assessed similarly to previous populations from Zielonka Forest complex on the base of the collected discs. This population represented the colonization stage and was located in a size-limited forest patch with a total area of 8 ha and surrounded by an extensive agricultural landscape (Table 1). The nearest black cherry population with adult trees was ca. 1 km away in a Scots pine stand 11 ha in size.
| Table 1. Characteristics of four studied populations of Prunus serotina located in Pinus sylvestris stands. | |||||||||
| Population | ID | Location | Number of individuals sampled | Habitat type | Invasion stage | No of adults | Density (indv./m2) | Age | P. sylvestris stand age |
| Raszków | R | 51°42’37.89” N 17°44’32.12” E | 109 | agricultural | colonization (isolated stand) | 8 | 0.18 | 17 | 34 |
| Zielonka Forest | Z_1 | 52°30’26.17” N 17°07’08.52” E | 180 | forest | colonization | 3 | 0.68 | 20 | 34 |
| Zielonka Forest | Z_2 | 52°31’00.81” N 17°06’44.46” E | 446 | forest | established | 16 | 1.82 | 35 | 80 |
| Zielonka Forest | Z_3 | 52°31’01.44” N 17°06’57.01” E | 205 | forest | established | 16 | 0.84 | 35 | 146 |
We adopted a star-like sampling scheme in order to simultaneously estimate clonal structure and FSGS (Fig. 1). For this reason, four 60 × 1 m crisscross transects running along the directions N–S, E–W, NW–SE and NE–SW were randomly selected in each of the P. serotina populations in Spring 2014. In total, 940 individuals were sampled and their position was recorded – 180 individuals in population Z_1, 446 in Z_2, 205 in Z_3 and 109 in R. Also, the numbers and positions of flowering individuals, and densities in terms of numbers of individuals per square meter were noted (Table 1). In all populations, the number of basal resprouts was counted. However, due to the limited number of basal resprouts noted in population Z_1 (2 individuals, see Results and Table 3), the significance of difference in average number of basal resprouts was tested only between population Z_2 and Z_3 with a T-test.
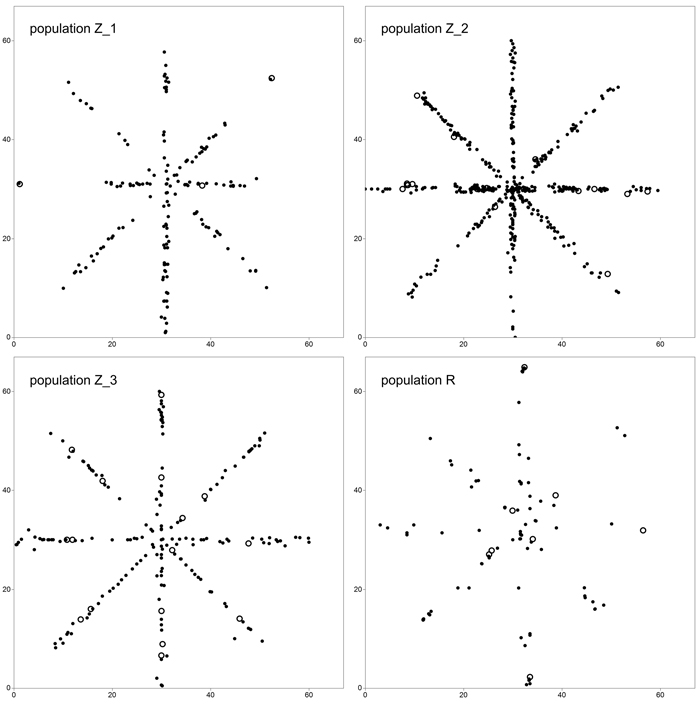
Fig. 1. Sampling scheme in study plots located in four analyzed populations of Prunus serotina. Open circles define location of mature (fruiting) trees. View larger in new window/tab
2.2 Genotyping
DNA was extracted from fresh plant tissue using the protocol of Dumolin et al. (1995). Genotyping was made with a set of eight nuclear microsatellite markers (nSSRs) which were originally generated for Prunus cerasus L. (sour cherry) and P. persica (L.) Batsch (peach) (Cipriani et al. 1999; Downey and Iezzoni 2000). Those markers were successfully used in previous studies in population genetics of P. serotina, which confirmed their high polymorphism (Pairon et al. 2008) (Table 2). Prunus serotina is a tetraploid and presents disomic mode of inheritance which was proven in controlled crosses (Pairon and Jacquemart 2005). The Polymerase Chains Reactions (PCR) were conducted in two PCR-Multiplex reactions using a Qiagen Multiplex PCR Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, with ca. 30 ng of template DNA and the thermal protocol used in Pairon et al. (2008). The PCR products were processed on a 3130 DNA Genetic Analyzer (Applied Biosystems) with the GeneScan 500-LIZ size standard. The software GeneMapper v.4.0 (Applied Biosystems) was used to score the genotypes.
| Table 2. Characteristics of eight microsatellite loci used. | |||||
| Locus | A | HO | HE | FIS | Null |
| UCD_CH14 | 17 | 0.761 | 0.782 | 0.027 | 0.023 |
| UDP96_005 | 6 | 0.482 | 0.584 | 0.175 | 0.070 |
| UDP98_025 | 9 | 0.723 | 0.757 | 0.045 | 0.027 |
| PCGA43 | 18 | 0.738 | 0.758 | 0.026 | 0.020 |
| PCHGMS3_1 | 20 | 0.639 | 0.813 | 0.214 | 0.090 |
| PCHGMS3_2 | 21 | 0.708 | 0.820 | 0.137 | 0.056 |
| PCHGSM2 | 18 | 0.642 | 0.674 | 0.047 | 0.022 |
| UDP98-405 | 12 | 0.474 | 0.564 | 0.161 | 0.062 |
| Average | 15.1 | 0.646 | 0.719 | 0.104 | 0.046 |
| A – number of alleles, HO – observed heterozygosity, HE – expected heterozygosity, FIS – inbreeding coefficient, Null – frequency of null alleles | |||||
2.3 Data analysis
The average numbers of alleles (A), observed (HO) and expected heterozygosities (HE), and inbreeding coefficients (FIS) were calculated for each population at genet level using GenAlEx v.6.2 (Peakall and Smouse 2006). The Hardy-Weinberg equilibrium (HWE) was tested in GenePop v. 4.0 using an exact test with 1000 iterations (Raymond and Rousset 1995). The frequency of null alleles (Null) was provided by INEst 1.0 (Chybicki and Burczyk 2009). Identification of genets and estimation of the probability that the repeated genotypes originate from independent reproduction events (psexHWE) (conservative mode including HWE departure) were done with GeneClone 2.0 (Arnaud-Haond and Belkhir 2006). Clonal diversity (R) of the population was estimated according to the formula R = (G–1) / (N–1), where G is the number of genets and N is the number of sampled individuals (ramets) (Dorken and Eckert 2001); value close to 1 denotes high input of generative reproduction i.e., high clonal diversity.
Parental relationships (genets only) were reconstructed using Colony 2.0.4.4, a maximum-likelihood parentage software (Jones and Wang 2010) in order to compare the sibship relationships among investigated populations with reference to the inferred FSGS. We used the following settings: monoecious species, inbreeding not present, diploid, polygamy for males and females, full-likelihood method, medium length run, medium precision and no updating allele frequencies. Locus-specific estimates of genotyping error rates per population were provided by INEst 1.0 (Chybicki and Burczyk 2009). The prior probability of finding a father or mother in the population was set to 0.5, but the prior information about sibship size was not given. Only relationships of full-sibs (FSs) and half-sibs (HSs) supported with a probability of >0.95 were scored.
Spatial structure may vary widely between populations, but comparing populations can be problematic because of differences in sampling design. To avoid this problem, for each population we used the same strategy during field sampling and analyzed FSGS using the same distance classes. Spatial auto-correlation analysis of kinship was conducted at genet level with SPAGeDi v. 1.2 (Hardy and Vekemans 2002). Average multilocus kinship coefficients (Fij) were computed (Loiselle et al. 1995) and averaged within each of the 10 distance classes: 5, 10, 15, 20, 25, 30, 40, 50, 60 and 65 m. Additionally, confidence intervals for the observed F(ij) were calculated as ±2SE, where SE is the standard error obtained after jacknifing over loci. The 95% confidence interval for each distance class was obtained from 10 000 permutations of the individual location. Average values of Fij were regressed on the logarithm of distance ln(dij) in order to obtain the regression slope, bF, which quantifies the extent of FSGS. The Sp statistic, developed by Vekemans and Hardy (2004), was computed according to the formula Sp = –bF / (1–F(1)), where F(1) refers to the mean kinship coefficient for individuals from the first distance class.
3 Results
3.1 Genetic diversity and population density
In total, we genotyped 940 individuals of black cherry. All used loci were polymorphic with varying numbers of alleles (A), from 6 to 21 with an average of 15.1 (Table 2). The frequency of null alleles (Null) ranged from 0.020 to 0.090, giving an average of 0.046 − a value frequently found in tree species. All loci presented significant deviations from HWE with FIS values ranging from 0.026 to 0.214 which indicates an excess of homozygotes.
Population statistics for all four investigated populations are given in Table 3. The highest average number of alleles per locus (A) was found in Z_3 (11.12), while the lowest was in the isolated population R (9.125). In all populations, a significant excess of homozygotes was observed. The highest FIS was noted in the isolated population R (0.137), and the lowest in population Z_2 (0.013).
| Table 3. Results of genetic structure, clonal diversity and pedigree analysis conducted in four populations of Prunus serotina. View in new window/tab. |
We noted different densities of black cherry in the analyzed populations (Table 1). The highest density had Z_2 (1.82 indv. /m2), while the two stands in the colonization stage presented lower densities, of which the isolated population R had the lowest (0.18 indv. /m2). In both the colonizing stage populations a considerably lower number of mature individuals was noted compared with two older populations (Table 1).
3.2 Clonality
In procedure of the genet identification, no mismatches were detected by GeneClone. The values of probabilities that the repeated genotype derives from distinct generative reproduction event (psexHWE) were reasonably low that supported the precision of the genet identification with only eight SSRs loci used (R: 4.95 ×10–125 – 6.02 × 10–4; Z_1: 9.92 ×10–48 – 1.98 × 10–10; Z_2: 1.34 ×10–16 – 3.74 × 10–9; Z_3: 7.09 ×10–15 – 2.15 × 10–12). The studied populations showed varied but generally low levels of clonal growth (Table 3). The highest clonal diversity (the proportion of all genets detected to the total number of ramets sampled) was noted in stand Z_3 (0.980), in which most of the sampled individuals were single-individual genets; the lowest clonal diversity was noted in the isolated stand R (0.713). In this population, the highest number of ramets per genets was noted (13). In the remaining populations, the input of clonality was low and two-individual genets prevailed (Table 3). We also noted differences in the number of basal resprouts. Generally, polycormic individuals prevailed in the older populations (Z_2 and Z_3), while being absent in the colonization stage, except for population Z_1, where only 2 such trees were found. The inclusion of basal resprouts increased the total level of clonality (Table 3). The average number of basal resprouts in population Z_3 was significantly higher than in Z_2 (3.18 vs. 2.32, respectively; p < 0.05).
3.3 Fine-scale genetic structure and pedigree reconstruction
The results of FSGS analyses in the four black cherry populations are presented in Table 4 and Fig. 2. Overall, we observed similar patterns across the investigated populations, with the highest significant positive relationship coefficient in the first distance interval (0–5 m), and decreasing continuously thereafter with increasing spatial distance (Table 4, Fig. 2). Closer inspection of the results revealed a difference between the populations from Zielonka Forest (Z_1–Z_3) and the isolated population R. Stronger FSGS as indicated by the over two-fold higher value of Sp was reported in the isolated population R in comparison to the three stands from Zielonka Forest for which the estimators of FSGS were, generally, at very similar levels. Also, the level of significant positive FSGS in terms of the pairwise kinship coefficient at the first distance class was highest in population R (Fij = 0.0346), but among populations from Zielonka Forest, the highest FSGS was in Z_3 (Fij = 0.0232). A negative significant FSGS was observed in populations Z_1 in the 40 m and 50 m and in Z_2 in the 30 m and 40 m distance classes.
| Table 4. Fine-scale genetic structure parameters for each population. | ||||
| Studied populations | Fij | bF | Sp | |
| R | Genets Ramets | 0.0346 (0.0075) 0.1568 (0.0239) | –0.01402*** (0.0033) –0.05107*** (0.0089) | 0.0145 0.0601 |
| Z_1 | Genets Ramets | 0.0143 (0.0030) 0.0195 (0.0040) | –0.00672*** (0.0011) –0.01010*** (0.0017) | 0.0068 0.0103 |
| Z_2 | Genets Ramets | 0.0119 (0.0011) 0.0120 (0.0011) | –0.00542*** (0.0006) –0.00558*** (0.0006) | 0.0054 0.0056 |
| Z_3 | Genets Ramets | 0.0232 (0.0059) 0.0258 (0.0057) | –0.00648*** (0.0017) –0.00515*** (0.0012) | 0.0066 0.0053 |
| Fij – the mean kinship coefficient between individuals in the first distance class (5 m), bF – slope of the regression of kinship coefficients on the logarithm of spatial distance, Sp – intensity of FSGS, *** – significant at 0.001, SE – in brackets. | ||||
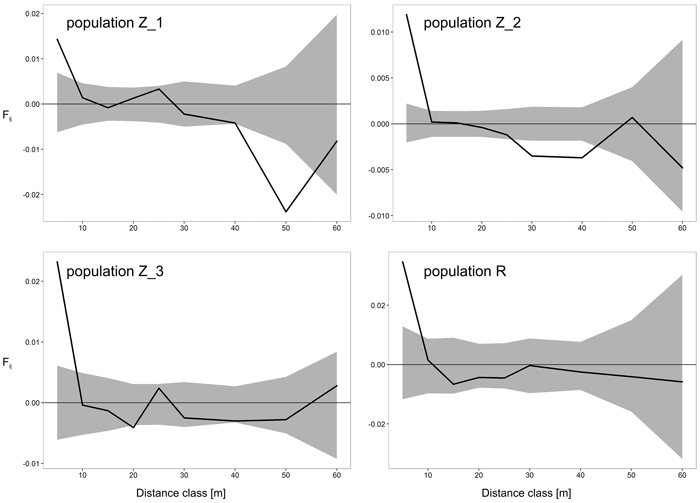
Fig. 2. Spatial correlograms for four analyzed Prunus serotina populations with mean pairwise kinship coefficients (Fij) of six distance classes for eight microsatellite loci. Upper and lower 95% confidence intervals for null hypothesis of random distribution of individuals are shaded. View larger in new window/tab
The results of pedigree reconstruction are presented in Table 3 as the percent of detected FSs pairs and HSs pairs to the total number of possible pair combinations. The analysis revealed the existence of relatives in all studied populations except for Z_1 where HSs were not detected. The highest proportion of HSs was in the isolated population R (5.39 %), while the highest proportion of FSs was in population Z_2 (almost 1%). Given the results on sibship structure in the three populations from Zielonka Forest the increase of HSs pairs was noticeable.
4 Discussion
In this work, we aimed to analyze the FSGS of populations of P. serotina, a successful invader of European forests. We traced changes in FSGS in populations at colonization and established stage of the invasive spread. There was an apparent difference in the levels of FSGS and relatedness in the two youngest populations of black cherry of which one was the isolated stand located in agricultural landscape (R) and the second (Z_1) was in continuous forest. In contrast, populations different with respect to the stage of the invasive spread (colonization vs. established) but located in the same continuous forest complex showed comparable levels of FSGS but varied relatedness. Thus, our results suggest that FSGS in black cherry may result from a complex set of demographic and ecological factors which are of varying importance during the process of invasive spread and will be discussed in detail below.
4.1 Colonization in agricultural landscape vs. forest complex
Our results show, that propagule source distance is the factor that may largely determine the process of invasive colonization in black cherry, as it was previously suggested in field observations (Knight et al. 2008; Jagodziński et al. 2015). Pairon et al. (2006) indicated that 83% of seeds of P. serotina fall in close vicinity of the mother tree − up to 5 m, and the remaining part of seed crop is dispersed by birds and can be transported up to 45 m. However, due to methodological constraints in that study, true long-distance dispersal episodes may not have been captured. Recently, based on indirect estimations Jagodziński et al. (2015) reported the maximum distance of seed dispersal for black cherry to be ca. 600 m. In our study, the isolation of the studied forest patch R allowed us to indirectly estimate long distance dispersal to be at least 1 km, because the closest population that could have been the source was located at that distance. Hence, P. serotina shows a stratified mode of seed dispersal, with a combination of short- and long-distance seed movements which refers to mixed type of seed dispersal (autochory – gravity and zoochory). Zoochory, which accounts for long-distance dispersal, promotes the species’ invasive spread by facilitating colonization events, similarly to the congeneric Prunus mahaleb L., which invades Argentine Pampas (Amodeo and Zalba 2013). In its native range, P. mahaleb was shown to disperse seeds at considerable distances, reaching over 900 m, but local seed movements dominate, similarly to P. serotina (García et al. 2007). The role of long distance dispersal is one of the focal points in studies on biological invasions, emphasizing the great invasive potential of frugivorous species (Von Der Lippe and Kowarik 2007; Vanhellemont et al. 2010; Pergl et al. 2011; Amodeo and Zalba 2013).
Gosper et al. (2005) showed that the structure of the landscape, i.e. barriers (e.g. low forest cover or fragmentation) and facilities (e.g. shrub cover within patches or perch trees) which affect connectivity among habitats within the landscape, shapes the dispersal pattern during invasive spread of the invasive species. Landscape features may affect the behavior of the dispersal agent and hence the seed dispersal effectiveness, as was proven in many studies (Santos et al. 1999; Figueroa-Esquivel et al. 2009; Schupp et al. 2010; Zapata et al. 2014; Lediuk et al. 2016). For example, the electricity pylons were showed to effectively facilitate the dispersal of P. serotina (Kurek et al. 2014). The investigations of Deckers et al. (2005) showed high directionality in the invasive colonization of black cherry in an agricultural landscape that was dependent on the spatial arrangement of hedgerow intersections. In their study, they showed that birds preferred moving along the hedgerow structures and not across the open space, which strongly shaped seed dispersion and further seedling occurrence. In our study, the isolated population R is a small forest patch located in an agricultural system that limits the passing of dispersers from one forest patch to another. Consequently, R received less colonizers than Z_1, which was surrounded by extensive forest habitat and numerous propagule sources. Although both the main dispersers of P. serotina seeds (Deckers et al. 2008) – Turdus merula L. (common blackbird) and Columba palumbus L. (common wood pigeon) show strong suburbanization (Evans et al. 2010), they are both primary forest bird species. Their behavior may be less effective for seed dispersal in the open space; thus, producing a considerable lag-phase in the colonization of isolated stands in agricultural landscape.
Black cherry is self-incompatible (SI); species with this mating system should present low or insignificant spatial structure (Vekemans and Hardy 2004; Hoebee et al. 2006). The value of the Sp statistic obtained in the isolated population R (0.0145) fell in the upper values previously noted for tree species with the SI system (Vekemans and Hardy 2004; Jankowska-Wroblewska et al. 2016) while in Z_1 (0.0068) fell within the lower values (Oddou-Muratorio and Klein 2008; Schueler et al. 2006). This shows that there is considerable variability in the spatial genetic structure of this species, which likely reflects the general complexity of demo-genetic factors involved in generating FSGS in plant populations (Jump et al. 2012; Moran and Clark 2012).
While long-distance seed dispersal promotes invasive spread into new sites and shapes the pattern of genetic differentiation among populations, short-distance dispersal determines the population dynamics and intra-population genetic structure (Jones et al. 2006; Hampe et al. 2010). Limited seed dispersal is mostly what drives the occurrence of significant FSGS in natural populations (Vekemans and Hardy 2004), and as indicated by Pairon et al. (2006), short-distance seed dispersal predominates in black cherry. Hence, limited dispersal would be the major driving force of the FSGS noted in studied populations. However, studies show that the number of founders, relatedness of initial colonists, effective density and other factors may have direct repercussions for FSGS (Jones et al. 2006). We noted a pronounced difference in FSGS between these two populations at colonization stage (Table 4). In the isolated population R we noted eight adult individuals which were probably the first founders, while only three such individuals were noted in population Z_1 (Table 1). This, at least theoretically, should foster greater FSGS in the latter, but we noted just the opposite: the strength of FSGS was 2-fold higher in R in comparison to Z_1. In our opinion, recruitment in R was mainly due to the local production of seeds by those eight parental trees, with limited seed immigration, while recruitment in Z_1 was primarily based on multiple seed sources located outside our mapped plots. Comparisons of sibship structure between these two populations (Table 3) seem to support our hypothesis. Pardini and Hamrick (2008) acknowledged the impact of the adjacently distributed adults on the dilution of FSGS in focal population. The long-lasting occurrence and hence abundance of black cherry in Zielonka Forest implies opportunities for seed immigration into our study plots due to seed shadow overlap, which is a factor preventing spatial structure (Hamrick et al. 1993). For example, Jones et al. (2006) argued that the low FSGS noted in a Pinus strobus L. population was due to a historical overlap of seed shadows, in comparison to the significant spatial structure reported in a newly established population of Q. rubra due to limited number of founding individuals. In a series of simulations, Moran and Clark (2012) confirmed that stronger FSGS coincides with a small number of within-population seed sources.
4.2 FSGS in colonization and established population
Studies exploring temporal changes in FSGS indicate that it is was diluting over time from seedlings to adults (Chung et al. 2003; Zhou and Chen 2010) and from initially colonized to fully established populations (Chung et al. 2007), although a reverse trend is also noted (Pardini and Hamrick 2008; Berens et al. 2014). The populations of black cherry from the Zielonka Forest complex representing different stages of the invasive spread presented very similar low intensities and extents of FSGS and varied sibship structures (Fig. 2, Table 3), which is contrary to our initial expectations. We predicted stronger FSGS in the colonizing population due to a residual founder effect vs. weaker or even insignificant FSGS in the established stage due to seed shadow overlap, opportunities for seed immigration from surroundings, and demographic thinning acting in the later stages. The homogenic FSGS across the two analyzed stages of invasive spread is likely due to the lack of an evident founder effect in the colonizing population Z_1 owing to an extensive base of propagules in the surroundings. Low kinship among individuals in the colonizing population Z_1 (none of them was HS and 0.17% were FS) supports the hypothesis of the lack of a founder effect.
The densities of seedlings and saplings were very similar in the colonizing population Z_1 and established Z_3 but in the second established stage population Z_2, the density was over two times higher. A demographic model of expansion described for P. mahaleb invading the grasslands of Argentina included lowest density in colonization stage and highest in the proliferation stage and lowering of the density in the mature population (Amodeo and Zalba 2013). As indicated the dendrochronological analysis, population Z_2 and Z_3 were founded at the same time but higher density of individuals in Z_2 suggests a proliferation stage which is characterized by massive fruit production (Amodeo and Zalba 2013). In our opinion, the difference between those two populations in their demographic stages can be best explained by competition with Q. robur. Population Z_2 is pure Scots pine stand while in Z_3 80-year-old oak constitutes a second forest-floor making up ca. 7% of the stand. Studies on invasion patterns in P. serotina consistently report its high colonization effectiveness in Scots pine stands, or generally in coniferous stands and young forests, in contrast to lower effectiveness in fully developed forests and non-coniferous stands (Chabrerie et al. 2007a, 2007b; Vanhellemont et al. 2009; Vanhellemont et al. 2010; Jagodziński et al. 2015). This trend reflects the divergent light conditions in these stands and light demands of black cherry, which attains highest growth, reproduction and probability of colonization in good light conditions (Closset-Kopp et al. 2007; Knight et al. 2008; Closset-Kopp et al. 2011; Jagodziński et al. 2015).
4.3 Clonality
We identified the presence of two forms of vegetative regeneration in the studied populations of black cherry: basal (stump) and root sucker regeneration. Both were reported previously for this species (Acuilar 1975; Closset-Kopp et al. 2007). Contrary to our initial expectations and previous reports for black cherry (Closset-Kopp et al. 2007), we noted rather low overall clonality due to root suckers. The high values of the parameter R that characterizes the level of clonal structure clearly indicate generative reproduction as the main source of new recruitments. Some invasive species even show a transition into greater vegetative reproduction in an adventive range (Castro et al. 2016), but this is not the case for black cherry. Considerable input of clonal individuals due to root suckers was noted only for the isolated population R, where the biggest genet of 13 ramets was detected. Existing data from the invasive range indicate that 10–13.7% of individuals are root suckers (Closset-Kopp et al. 2007; Halarewicz 2011; Jagodziński et al. 2015). It is also possible that our sampling strategy did not allow all ramets to be detected in study plots. Nevertheless, the multiplications of genotypes by clonal reproduction significantly reinforced the level of FSGS in studied populations, which was in accordance with our expectations and other studies that compared intensity of FSGS at genet and ramet level (Vaughan et al. 2007; Dering et al. 2016); this was especially noticeable in population R (Table 4). However, somewhat lower value of Sp at ramet level than at genet level noted in population Z_3, at first surprising, may only be interpreted in frame of the clonal architecture. A guerrilla type of clonal growth in which ramets of different genets intermingle may dissolve a FSGS in contrary to phalanx growth which may intensify it (Dodd et al. 2013; Dering et al. 2015).
The overall clonality level increased if basal resprouts were included (Table 3), and this form of clonal growth seems to be more important than root suckering. Closset-Kopp et al. (2007) reported basal resprouts to be as high as 51.7% of individuals and prevailed mainly among young seedlings irrespective of light conditions. In contrast, our field observations indicate that basal resprouts are more frequent in older seedlings and saplings and do not attain such high frequencies (Table 3). In the native range, Auclair (1975) claimed that black cherry is one of the most dynamically resprouting tree species. According to the author, in black cherry increased competition from overstorey oak trees that leads to reduced species growth rates, induces regeneration from basal sprouts. We follow this interpretation because the highest frequency of basal resprouts was noted in population Z_3 in which the oak second forest-floor was present, which probably modifies the light conditions of the stand.
Our results indicate that vegetative regeneration is an important life-history trait in P. serotina and may contribute to its invasiveness. In P. serotina vegetative regeneration is viewed as strategy for long-term persistence in stochastic environments and sub-optimal light conditions (Auclair 1975; Closset-Kopp et al. 2007). Based on our results and field observations we conclude that clonality per se is not a strategy allowing colonization, efficient space takeover or outcompeting of other species as was proven to be the case in other invasive species capable of vast clonal spread (Hollingsworth and Bailey 2000; Kollmann et al. 2009; Budde et al. 2010; Castro et al. 2016). We rather consider that for black cherry, clonality mediates the transition from durable maintenance in a recipient habitat with sub-optimal conditions (low light availability) to the abundant seed production phase, which is one of the main contributors to the great invasiveness of black cherry. In favorable conditions, e.g. after opening of the forest canopy, each of the basal resprouts may start and/or increase the production of flowers and fruits, triggering invasive spread by the greater reproductive output of individuals (Vallejo-Marín et al. 2010). At the population scale, an increased production of seeds would positively affect the frugivore service that is crucial for this species to spread into new territories. Therefore, by efficient long-term supporting of a seedling bank, clonality emerges as a component of the ‘sit-and-wait’ strategy in black cherry and recognized as factor for its successful invasion (Closset-Kopp et al. 2007). The development and maintenance of a seedling/sapling bank was also suggested to be an important factor for the invasiveness of Acer platanoides L. and Sorbus acuparia L. in their non-native ranges (Martin and Marks 2006; Lediuk et al. 2016).
5 Conclusions
It was previously shown that P. serotina is able to shuffle between different demographic strategies depending on the prevailing conditions in the recipient habitat in order to efficiently invade and persist in a forest ecosystem, and possesses some of the traits that match the profiles of many other successful invaders (Closset-Kopp et al. 2007; Robakowski and Bielinis 2011). Our study is another contribution confirming the great invasive potential of this species in Scots pine forests. In this study, we showed that black cherry’s invasive spread is strongly determined by propagule availability and landscape features that modify the course of the invasive spread with respect to rate and the population-level distribution of genetic information. Consequently, in contrast to stands surrounded by an extensive base of propagules, colonization of isolated stands may involve the emerging of a lag-phase and clustering of relatives due to the founder effect. A lag-phase is potentially the best opportunity for efficient management of the species. Another practical hint that emerges from our study is that the forest management of should focus on maintaining the closed canopy in the forest stands in order to decrease risk of the invasion and further expansion of the invader. It means also the need of transformation of pure Scots pine stands into mixed stands as previously suggested Jagodzinski et al. (2015). In population Z_3, the presence of the oak in the second forest-floor of the oldest Scots pine stands likely reduced survivorship of young individuals and resulted in lower density. To our surprise, clonality doesn’t seem to be an underlying factor of black cherry invasiveness, but may efficiently support a seedling/sapling bank, which is considered to be a crucial factor in species’ invasive strategy.
Significant FSGS in species with the SI system, which includes the black cherry, may negatively affect future population persistence and performance because the Allele effect may negatively affect mean fitness and thus lower the probability of invader establishment (Willi 2005). Our quantification of FSGS in this study was based on neutral genetic markers, and studies show that these may not follow the spatial structure of the S-RNase locus (Jankowska-Wroblewska et al. 2016; Jolivet et al. 2010). Hence, it seems that in invasive species with the SI system, including the S-RNase locus into investigations would provide more comprehensive insight into the mechanism governing the spatial distribution of genetic information and thus directions of population process relevant for invasive spread. Additionally, the strong FSGS reported here for the isolated population R raises the interesting possibility of a partial breakdown of the SI system in P. serotina in its invasive range. This would allow avoiding the negative consequences of spatial clustering of relatives and could undoubtedly be a significant factor for species invasiveness. In fact, transitions from self-incompatibility to self-compatibility were described in the congeneric Prunus cerasus L., where it was based on the accumulation of nonfunctional S-haplotypes (Hauck et al. 2006).
Finally, broader population studies of the temporal changes of FSGS are needed. These should incorporate data on mating patterns and detailed information on SI diversity, seed- and pollen-mediated gene flow rate and demography. This would allow deeper understanding of the process of invasive spread of black cherry and its influencing factors. The role of isolated populations seems to be especially interesting – are they important for accelerating successful range expansion (Lawson Handley et al. 2011), or are they just wasted chances?
Acknowledgements
Authors thank M. Łuczak and R. Dering for technical assistance in laboratory and field works, and I.J Chybicki for support with statistics. We are also grateful to the Wronczyn Forest District and Łopuchówko Forest District for possibility to conduct the field research at their locations. This work was financed by Institute of Dendrology, Polish Academy of Sciences (Statutory Founds).
References
Aerts R., Ewald M., Nicolas M., Piat J., Skowronek S., Lenoir J., Hattab T., Garzón-López C.X., Feilhauer H., Schmidtlein S., Rocchini D., Decocq G., Somers B., Van De Kerchove R., Denef K., Honnay O. (2017). Invasion by the alien tree Prunus serotina alters ecosystem functions in a temperate deciduous forest. Frontiers in Plant Science 8: 179. 11 p. https://doi.org/10.3389/fpls.2017.00179.
Águas A., Larcombe M., Matias H., Deus E, Pott B.M., Rego F.C., Silva J.S. (2017). Understanding the naturalization of Eucalyptus globulus in Portugal: a comparison with Australian plantations. European Journal of Forest Research 136: 433–446. https://doi.org/10.1007/s10342-017-1043-6.
Amodeo M.R., Zalba S.M. (2013). Wild cherries invading natural grasslands: unravelling colonization history from population structure and spatial patterns. Plant Ecology 214: 1299–1307. https://doi.org/10.1007/s11258-013-0252-4.
Arnaud-Haond S., Belkhir K. (2006). GENECLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Molecular Ecology Notes 7: 15–17. https://doi.org/10.1111/j.1471-8286.2006.01522.x.
Auclair A.N. (1975). Sprouting response in Prunus serotina Erhr.: multi-variate analysis of site, forest structure and growth rate relationships. The American Midland Naturalist 94: 72–87. https://doi.org/10.2307/2424539.
Barnaud A., Kalwij J.M., Berthouly-Salazar C., McGeoch M.A., Jansen van Vuuren B. (2013). Are road verges corridors for weed invasion? Insights from the fine-scale spatial genetic structure of Raphanus raphanistrum. Weed Research 53: 362–369. https://doi.org/10.1111/wre.12033.
Berens D.G., Braun C., González-Martínez S.C., Griebeler E.M., Nathan R., Böhning-Gaese K. (2014). Fine-scale spatial genetic dynamics over the life cycle of the tropical tree Prunus africana. Heredity 11: 401–407. https://doi.org/10.1038/hdy.2014.40.
Blackburn T.M., Pyšek P., Bacher S., Carlton J.T., Duncan R.P., Jarošík V., Wilson J.R.U., Richardson D.M. (2011). A proposed unified framework for biological invasions. Trends in Ecology and Evolution 26: 333–339. https://doi.org/10.1016/j.tree.2011.03.023.
Blackburn T.M., Lockwood J.L., Cassey P. (2015). The influence of numbers on invasion success. Molecular Ecology 24: 1942–1953. https://doi.org/10.1111/mec.13075.
Bock D.G., Caseys C., Cousens R.D., Hahn M.A., Heredia S.M., Hübner S., Turner K.G., Whitney K.D., Rieseberg L.H. (2015). What we still don’t know about invasion genetics. Molecular Ecology 24: 2277–2297. https://doi.org/ 10.1111/mec.13032.
Bravo-Monasterio P., Pauchard A., Fajardo A. (2016). Pinus contorta invasion into treeless steppe reduces species richness and alters species traits of the local community. Biological Invasions 18: 1883–1894. https://doi.org/10.1007/s10530-016-1131-4.
Budde K.B., Gallo L., Marchelli P., Mosner E., Liepelt S., Ziegenhagen B., Leyer I. (2010). Wide spread invasion without sexual reproduction? A case study on European willows in Patagonia, Argentina. Biological Invasions 13: 45–54. https://doi.org/10.1007/s10530-010-9785-9.
Capinha C., Essl F., Seebens H., Moser D., Pereira H.M. (2015). The dispersal of alien species redefines biogeography in the Anthropocene. Science 348: 1248–1251. https://doi.org/10.1126/science.aaa8913.
Castro S., Castro M., Ferrero V., Costa J., Tavares D., Navarro L., Loureiro J. (2016). Invasion fosters change: independent evolutionary shifts in reproductive traits after Oxalis pes-caprae L. introduction. Frontiers in Plant Science 7: 784. https://doi.org/10.3389/fpls.2016.00874.
Chabrerie O., Roulier F., Hoeblich H., Sebert-Cuvillier E., Closset-Kopp D., Leblanc I., Jaminon J., Decocq G. (2007a). Defining patch mosaic functional types to predict invasion patterns in a forest landscape. Ecological Applications 17: 464–481. https://doi.org/10.1890/06-0614.
Chabrerie O., Verheyen K., Saguez R., Decocq G. (2007b). Disentangling relationships between habitat conditions, disturbance history, plant diversity, and American black cherry (Prunus serotina Ehrh.) invasion in a European temperate forest: Invasion-disturbance-ecosystem interactions. Diversity and Distribution 14: 204–212. https://doi.org/10.1111/j.1472-4642.2007.00453.x.
Chung M.Y., Epperson B.K., Gi Chung M. (2003). Genetic structure of age classes in Camellia japonica (Theaceae). Evolution 57: 62–73. https://doi.org/10.1111/j.0014-3820.2003.tb00216.x.
Chung M.Y., Nason J.D., Chung M.G. (2007). Effects of population succession on demographic and genetic processes: predictions and tests in the daylily Hemerocallis thunbergii (Liliaceae). Molecular Ecology 16: 2816–2829. https://doi.org/10.1111/j.1365-294X.2007.03361.x.
Chybicki I.J., Burczyk J. (2009). Simultaneous estimation of null alleles and inbreeding coefficients. Journal of Heredity 100: 106-113. https://doi.org/10.1093/jhered/esn088.
Chybicki I.J., Oleksa A., Burczyk J. (2011). Increased inbreeding and strong kinship structure in Taxus baccata estimated from both AFLP and SSR data. Heredity 107: 589–600. https://doi.org/10.1038/hdy.2011.51.
Cipriani G., Lot G., Huang W.-G., Marrazzo M.T., Peterlunger E., Testolin R. (1999). AC/GT and AG/CT microsatellite repeats in peach [Prunus persica (L) Batsch]: isolation, characterisation and cross-species amplification in Prunus. Theoretical and Applied Genetics 99: 65–72. https://doi.org/10.1007/s001220051209.
Closset-Kopp D., Chabrerie O., Valentin B., Delachapelle H., Decocq G. (2007). When Oskar meets Alice: does a lack of trade-off in r/K-strategies make Prunus serotina a successful invader of European forests? Forest Ecology and Management 247: 120–130. https://doi.org/10.1016/j.foreco.2007.04.023.
Closset-Kopp D., Saguez R., Decocq G. (2011). Differential growth patterns and fitness may explain contrasted performances of the invasive Prunus serotina in its exotic range. Biological Invasions 13: 1341–1355. https://doi.org/10.1007/s10530-010-9893-6.
Craven D., Thakur M.P., Cameron E.K., Frelich L.E., Beauséjour Blair R.B., Blossey B., Burtis J., Choi A., Dávalos A., Fahey T.J., Fisichelli N.A., Gibson K., Handa T.I., Hopfenserger K., Loss S.R., Nuzzo V., Maerz J.C., Sackett T., Scharenbroch B.C., Smith S.M., Vellend M., Umerk L.G., Eisenhauer N. (2017). The unseen invaders: introduced earthworms as drivers of change in plant communities in North American forests (a meta-analysis). Global Change Biology 23: 1065–1074. https://doi.org/10.1111/gcb.13446.
Crowley S.L., Hinchliffe S., McDonald R.A. (2017). Invasive species management will benefit from social impact assessment. Journal of Applied Ecology 54: 351–357. https://doi.org/10.1111/1365-2664.12817.
DAISIE European Invasive Alien Species Gateway (2008). Prunus serotina. http://www.europe-aliens.org/speciesFactsheet.do?speciesId=13913. [Cited 16th July 2018].
Deckers B., Verheyen K., Hermy M., Muys B. (2005). Effects of landscape structure on the invasive spread of black cherry Prunus serotina in an agricultural landscape in Flanders, Belgium. Ecography 28: 99–109. https://doi.org/10.1111/j.0906-7590.2005.04054.x.
Deckers B., Verheyen K., Vanhellemont M., Maddens E., Muys B., Hermy M. (2008). Impact of avian frugivores on dispersal and recruitment of the invasive Prunus serotina in an agricultural landscape. Biological Invasions 10: 717–727. https://doi.org/10.1007/s10530-007-9164-3.
Dering M., Chybicki I.J., Rączka G. (2015). Clonality as a driver of spatial genetic structure in populations of clonal tree species. Journal of Plant Research 128: 731–745. https://doi.org/10.1007/s10265-015-0742-7.
Dering M., Rączka G., Szmyt J. (2016). Sex-specific pattern of spatial genetic structure in dioecious and clonal tree. species, Populus alba L. Tree Genetics and Genomes 12: 70. https://doi.org/10.1007/s11295-016-1028-5.
Dorken M.E., Eckert C.G. (2001). Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). Journal of Ecology 89: 339–350. https://doi.org/10.1046/j.1365-2745.2001.00558.x.
Downey S.L., Iezzoni A.F. (2000). Polymorphic DNA markers in black cherry (Prunus serotina) are identified using sequences from sweet cherry, peach, and sour cherry. Journal of the American Society of Horticulture Science 125: 76–80.
Dumolin S., Demesure B., Petit R.J. (1995). Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theoretical and Applied Genetics 91: 1253–1256. https://doi.org/10.1007/BF00220937.
Dylewski Ł., Kurek P., Wiatrowska B., Jerzak L., Tryjanowski P. (2017). Man-made perching sites – electricity pylons accelerate fleshy-fruited plants succession in farmlands. Flora 231: 51–56. https://doi.org/10.1016/j.flora.2017.04.004.
Evans K.L., Hatchwell B.J., Parnellm M., Gaston K. (2010). A conceptual framework for the colonisation of urban areas: the blackbird Turdus merula as a case study. Biological Reviews 85: 643–667. https://doi.org/10.1111/j.1469-185X.2010.00121.x.
Figueroa-Esquivel E., Puebla-Olivares F., Godínez-Álvarez H., Núñez-Farfán J. (2009). Seed dispersal effectiveness by understory birds on Dendropanax arboreus in a fragmented landscape. Biodiversity Conservation 18: 3357–3365. https://doi.org/10.1007/s10531-009-9645-z.
García C., Jordano P., Godoy J.A. (2007). Contemporary pollen and seed dispersal in a Prunus mahaleb population: patterns in distance and direction. Molecular Ecology 16:1947–1955. https://doi.org/10.1111/j.1365-294X.2006.03126.x.
Gaskin J.F., Schwarzländer M., Grevstad F.S., Haverhals M.A., Bourchier R.S., Miller T.W. (2014). Extreme differences in population structure and genetic diversity for three invasive congeners: knotweeds in western North America. Biological Invasions 16: 2127–2136. https://doi.org/10.1007/s10530-014-0652-y.
Gosper C.R., Stansbury C.D., Vivian-Smith G. (2005). Seed dispersal of fleshy-fruited invasive plants by birds: contributing factors and management options. Diversity and Distributions 11: 549–558. https://doi.org/10.1111/j.1366-9516.2005.00195.x.
Halarewicz A. (2011). Regeneration of black cherry (Prunus serotina Ehrh.) in coniferous forest communities. Sylwan 155: 530–534.
Halarewicz A., Żołnierz L. (2014). Changes in the understorey of mixed coniferous forest plant communities dominated by the American black cherry (Prunus serotina Ehrh.). Forest Ecology and Management 313: 91–97. https://doi.org/10.1016/j.foreco.2013.11.006.
Hampe A., El Masri L., Petit R.J. (2010). Origin of spatial genetic structure in an expanding oak population. Molecular Ecology 19: 459–471. https://doi.org/10.1111/j.1365-294X.2009.04492.x.
Hamrick J.L., Murawski D.A., Nason J.D. (1993). The influence of seed dispersal mechanisms on the genetic structure of tropical tree populations. Vegetatio 107: 281–297. https://doi.org/10.1007/BF00052230.
Hardy O., Vekemans X. (2002). SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Resources 2: 618–620. https://doi.org/10.1046/j.1471-8286.2002.00305.x.
Hauck N.R., Yamane H., Tao R., Iezzoni A.F. (2006). Accumulation of nonfunctional S-haplotypes results in the breakdown of gametophytic self-incompatibility in tetraploid Prunus. Genetics 172: 1191–1198. https://doi.org/10.1534/genetics.105.049395.
Herrera C.M., Jordano P., Lopez-Soria L., Amat J.A. (1994). Recruitment of a mast-fruiting, bird-dispersed tree: bridging frugivore activity and seedling establishment. Ecological Monographs 64: 315–344. https://doi.org/10.2307/2937165.
Hoebee S.E., Menn C., Rotach P., Finkeldey R., Holderegger R. (2006). Spatial genetic structure of Sorbus torminalis: the extent of clonal reproduction in natural stands of a rare tree species with a scattered distribution. Forest Ecology and Management 226: 1–8. https://doi.org/10.1016/j.foreco.2005.12.024.
Hollingsworth M.L., Bailey J.P. (2000). Evidence for massive clonal growth in the invasive weed Fallopia japonica (Japanese Knotweed). Botanical Journal of Linnean Society 133: 463–472. https://doi.org/10.1111/j.1095-8339.2000.tb01589.x.
Holmes R. (1994). Dendrochronology program library. User’s manual. University of Arizona, Tucson.
Hulme P.E. (2017). Climate change and biological invasions: evidence, expectations, and response options. Biological Reviews 92: 1297–1313. https://doi.org/10.1111/brv.12282.
Jagodziński A.M., Dyderski M.K., Rawlik M., Banaszczak P. (2015). Plantation of coniferous trees modifies risk and size of Padus serotina (Ehrh.) Borkh. invasion – evidence from a Rogów Arboretum case study. Forest Ecology and Management 357: 84–94. https://doi.org/10.1016/j.foreco.2015.08.011.
Jankowska-Wroblewska S., Warmbier J., Burczyk J. (2016). Spatial genetic structure within populations of Sorbus torminalis (L.) Crantz: comparative analysis of the self-incompatibility locus and nuclear microsatellites. Acta Biologica Cracoviense Series Botanica 58: 7–17. https://doi.org/10.1515/abcsb-2016-0011.
Jolivet C., Höltke A.M., Liesebach H., Steiner W., Degen B. (2010). Spatial genetic structure in wild cherry (Prunus avium L.): I. variation among natural populations of different density. Tree Genetics and Genomes 7: 271–283. https://doi.org/10.1007/s11295-010-0330-x.
Jones F.A., Hamrick J.L., Peterson C.J., Squiers E.R. (2006). Inferring colonization history from analyses of spatial genetic structure within populations of Pinus strobus and Quercus rubra. Molecular Ecology 15: 851–861. https://doi.org/10.1111/j.1365-294X.2005.02830.x.
Jones O.R., Wang J. (2010). COLONY: a program for parentage and sibship inference from multilocus genotype data. Molecular Ecology Resources 10: 551–555. https://doi.org/10.1111/j.1755-0998.2009.02787.x.
Jump A.S., Rico L., Coll M., Peñuelas J. (2012). Wide variation in spatial genetic structure between natural populations of the European beech (Fagus sylvatica) and its implications for SGS comparability. Heredity 108: 633–639. https://doi.org/10.1038/hdy.2012.1.
Karolewski P., Jagodziński A.M., Giertych M.J., Łukowski A., Baraniak E., Oleksyn J. (2014). Invasive Prunus serotina – a new host for Yponomeuta evonymellus (Lepidoptera: Yponomeutidae)? European Journal of Entomology 111: 227–236. https://doi.org/10.14411/eje.2014.026.
Kelager A., Pedersen J.S., Bruun H.H. (2013). Multiple introductions and no loss of genetic diversity: invasion history of Japanese Rose, Rosa rugosa, in Europe. Biological Invasions 15: 1125–1141. https://doi.org/10.1007/s10530-012-0356-0.
Knight K.S., Oleksyn J., Jagodzinski A.M., Reich P.B., Kasprowicz M. (2008). Overstorey tree species regulate colonization by native and exotic plants: a source of positive relationships between understorey diversity and invasibility. Diversity and Distributions 14: 666–675. https://doi.org/10.1111/j.1472-4642.2008.00468.x.
Kollmann J., Jørgensen R.H., Roelsgaard J., Skov-Petersen H. (2009). Establishment and clonal spread of the alien shrub Rosa rugosa in coastal dune – a method for reconstructing and predicting invasion patterns. Landscape Urban Planning 93: 194–200. https://doi.org/10.1016/j.landurbplan.2009.07.006.
Kurek P., Sparks T.H., Tryjanowski P. (2015). Electricity pylons may be potential foci for the invasion of black cherry Prunus serotina in intensive farmland. Acta Oecologica 62: 40–44. https://doi.org/10.1016/j.actao.2014.11.005.
Lawson Handley L.-J., Estoup A., Evans D.M., Thomas C.E. Lombaert E., Facon B., Aebi A., Roy H.E. (2011). Ecological genetics of invasive alien species. BioControl 56: 409–428. https://doi.org/10.1007/s10526-011-9386-2.
Lediuk K.D., Damascos M.A., Puntieri J.G., Curth M. de T. (2016). Population dynamics of an invasive tree, Sorbus aucuparia, in the understory of a Patagonian forest. Plant Ecology 217: 899–911. https://doi.org/10.1007/s11258-016-0615-8.
Liu J., Dong M., Miao S.L., Li Z.Y., Song M.H., Wang R.Q. (2006). Invasive alien plants in China: role of clonality and geographical origin. Biological Invasions 8: 1461–1470. https://doi.org/10.1007/s10530-005-5838-x.
Loiselle B.A., Sork V.L., Nason J., Graham C. (1995). Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). American Journal of Botany 82: 1420–1425. https://doi.org/10.1002/j.1537-2197.1995.tb12679.x.
Lorenz K., Preston C.M., Krumrei S., Feger K.-H. (2004). Decomposition of needle/leaf litter from Scots pine, black cherry, common oak and European beech at a conurbation forest site. European Journal of Forest Research 123: 177–188. https://doi.org/10.1007/s10342-004-0025-7.
Martin P.H., Marks P.L. (2006). Intact forests provide only weak resistance to a shade-tolerant invasive Norway maple (Acer platanoides L.). Journal of Ecology 94: 1070–1079. https://doi.org/10.1111/j.1365-2745.2006.01159.x.
Moran E.V., Clark J.S. (2012). Between-site differences in the scale of dispersal and gene flow in red oak. PLoS ONE 7: e36492. https://doi.org/10.1371/journal.pone.0036492.
Mostert E., Gaertner M., Holmes P.M., Rebelo A.G., Richardson D.M. (2017). Impacts of invasive alien trees on threatened lowland vegetation types in the Cape Floristic Region, South Africa. South African Journal of Botany 108: 209–222. https://doi.org/10.1016/j.sajb.2016.10.014.
Oddou-Muratorio S., Klein E.K. (2008). Comparing direct vs. indirect estimates of gene flow within a population of a scattered tree species. Molecular Ecology 17: 2743–2754. https://doi.org/10.1111/j.1365-294X.2008.03783.x.
Pairon M., Jonard M., Jacquemart A.-L. (2006). Modeling seed dispersal of black cherry, an invasive forest tree: how microsatellites may help? Canadian Journal of Forest Research 36: 1385–1394. https://doi.org/10.1139/x06-018.
Pairon M., Jacquemart A-L., Potter D. (2008). Detection and characterization of genome-specific microsatellite markers in the allotetraploid Prunus serotina. Journal of the American Society of Horticulture Sciences 133: 390–395.
Pairon M., Petitpierre B., Campbell M., Guisan A., Broennimann O., Baret P.V., Jacquemart A.-L., Besnard G. (2010). Multiple introductions boosted genetic diversity in the invasive range of black cherry (Prunus serotina; Rosaceae). Annals of Botany 105: 881–890. https://doi.org/10.1093/aob/mcq065.
Pardini E.A., Hamrick J.L. (2008). Inferring recruitment history from spatial genetic structure within populations of the colonizing tree Albizia julibrissin (Fabaceae). Molecular Ecology 17: 2865–2879. https://doi.org/10.1111/j.1365-294X.2008.03807.x.
Peakall R., Smouse P.E. (2006). genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6: 288–295. https://doi.org/10.1093/bioinformatics/bts460.
Pergl J., Müllerová J., Perglová I., Herben T., Pyšek P. (2011). The role of long-distance seed dispersal in the local population dynamics of an invasive plant species. Diversity and Distributions 17: 725–738. https://doi.org/10.1111/j.1472-4642.2011.00771.x.
Petitpierre B., Pairon M., Broennimann O., Jacquemart A.L., Guisan A., Besnard G. (2009). Plastid DNA variation in Prunus serotina var. serotina (Rosaceae), a North American tree invading Europe. European Journal of Forest Research 128: 431–436. https://doi.org/10.1007/s10342-009-0287-1.
Pyšek P., Richardson D.M. (2008). Traits associated with invasiveness in alien plants: where do we stand? In: Netwing W. (ed.). Biological invasions, ecological studies 193. Springer-Verlag, Berlin & Heidelberg. p. 97–125.
Rafajlović M., Kleihans D., Gulliksson C., Fries J., Johansson D., Ardehed A., Sundqvist L., Pereyra R.T., Mehling B., Jonsson P.R., Johannesson K. (2017). Neutral processes forming large clones during colonization of new areas. Journal of Evolutionary Biology 30: 1544 – 1560. https://doi.org/10.1111/jeb.13124.
Raymond M., Rousset F. (1995). An exact test for population differentiation. Evolution 49: 1280. https://doi.org/10.2307/2410454.
Richardson D.M., Pyšek P. (2006). Plant invasions: merging the concepts of species invasiveness and community invasibility. Progress in Physical Geography: Earth and Environment 30: 409–431. https://doi.org/10.1191/0309133306pp490pr.
Riepšas E., Straigytė L. (2008). Invasiveness and ecological effects of red oak (Quercus rubra L.) in Lithuanian forests. Baltic Forestry 14: 22–130.
Robakowski P., Bielinis E. (2011). Competition between sessile oak (Quercus petrea) and black cherry (Padus serotina): dynamics of seedlings growth. Polish Journal of Ecology 59: 297–306.
Sakai A.K., Allendorf F.W., Holt J.S., Lodge D.M., Molofsky J., With K.A., Baughman S., Cabin R.J., Cohen J.E., Ellstrand N.C., O’Nell P., Parker I.M., Thompson J.N., Weller S.W. (2001). The population biology of invasive species. Annual Reviews of Ecology and Systematics 32: 305–332. https://doi.org/10.1146/annurev.ecolsys.32.081501.114037.
Santos T., Telleria J.L., Virgos E. (1999). Dispersal of Spanish juniper Juniperus thurifera by birds and mammals in a fragmented landscape. Ecography 22: 193–204. https://doi.org/10.1111/j.1600-0587.1999.tb00468.x.
Schueler S., Tusch A., Scholz F. (2006). Comparative analysis of the within-population genetic structure in wild cherry (Prunus avium L.) at the self-incompatibility locus and nuclear microsatellites. Molecular Ecology 15: 3231–3243. https://doi.org/10.1111/j.1365-294X.2006.03029.x.
Schupp E.W., Jordano P., Gómez J.M. (2010). Seed dispersal effectiveness revisited: a conceptual review. New Phytologist 188: 333–353. https://doi.org/10.1111/j.1469-8137.2010.03402.x.
Selvi F., Carrari E., Coppi A. (2016). Impact of pine invasion on the taxonomic and phylogenetic diversity of a relict Mediterranean forest ecosystem. Forest Ecology and Management 367: 1–11. https://doi.org/10.1016/j.foreco.2016.02.013.
Sloop C.M., Ayres D.R., Strong D.R. (2011). Spatial and temporal genetic structure in a hybrid cordgrass invasion. Heredity 106: 547–556. https://doi.org/10.1038/hdy.2010.63.
Taylor K.T., Maxwell B.D., McWethy D.B., Pauchard A., Nuñez M.A., Whitlock C. (2017). Pinus contorta invasions increase wildfire fuel loads and may create a positive feedback with fire. Ecology 98: 678–687. https://doi.org/10.1002/ecy.1673.
Travis S.E., Marburger J.J., Windels S.K., Kubátová B. (2011). Clonal structure of invasive Cattail (Typhaceae) stands in the upper Midwest Region of the US. Wetlands 31: 221–228. https://doi.org/10.1007/s13157-010-0142-7.
Tyler T., Karlsson T., Milberg P., Sahlin U., Sundberg S. (2015). Invasive plant species in the Swedish flora: developing criteria and definitions and assessing the invasiveness of individual taxa. Nordic Journal of Botany 33: 300–317. https://doi.org/10.1111/njb.00773.
Vallejo-Marín M., Dorken M.E., Barrett S.C.H. (2010). The ecological and evolutionary consequences of clonality for plant mating. Annual Reviews in Ecology, Evolution and Systematics 41: 193–213. https://doi.org/10.1146/annurev.ecolsys.110308.120258.
Vanhellemont M., Verheyn K., De Keersmaeker L., Vandekerkhove K., Hermy M. (2009). Does Prunus serotina act as an aggressive invader in areas with a low propagule pressure? Biological Invasions 11: 1451–1462. https://doi.org/10.1007/s10530-008-9353-8.
Vanhellemont M., Wauters L., Baeten L., Bijlsma R.-J., Frenne P.D., Hermy M., Verheyen K. (2010). Prunus serotina unleashed: invader dominance after 70 years of forest development. Biological Invasions 12: 1113–1124. https://doi.org/10.1007/s10530-009-9529-x.
Van Wilgen B.W., Reyers B., Le Maitre D.C., Richardson D.M., Schonegevel L. (2008). A biome-scale assessment of the impact of invasive alien plants on ecosystem services in South Africa. Journal of Environmental Management 89: 336–349. https://doi.org/10.1016/j.jenvman.2007.06.015 015.
Vekemans X., Hardy O.J. (2004). New insights from fine-scale spatial genetic structure analyses in plant populations. Molecular Ecology 13: 921–935. https://doi.org/10.1046/j.1365-294X.2004.02076.x.
Von Der Lippe M., Kowarik I. (2007). Long-distance dispersal of plants by vehicles as a driver of plant invasions. Conservation Biology 21: 986–996. https://doi.org/10.1111/j.1523-1739.2007.00722.x.
Willi Y. (2005). A threefold genetic Allee effect: population size affects cross-compatibility, inbreeding depression and drift load in the self-incompatible Ranunculus reptans. Genetics 169: 2255–2265. https://doi.org/10.1534/genetics.104.034553.
Woziwoda B., Kopeć D., Witkowski J. (2014). The negative impact of intentionally introduced Quercus rubra L. on a forest community. Acta Societatis Botanicorum Poloniae 83: 39–49. https://doi.org/10.5586/asbp.2013.035.
Zapata V.M., Robledano F., Ramos V., Martínez-López V. (2014). Bird-mediated seed dispersal of fleshy fruits of Mediterranean shrubs in semiarid forest patches: the role of Pinus halepensis Miller trees as seed receptors. Plant Ecology 215: 337–1350. https://doi.org/10.1007/s11258-014-0391-2.
Zhou H.-P., Chen J. (2010). Spatial genetic structure in an understorey dioecious fig species: the roles of seed rain, seed and pollen-mediated gene flow, and local selection. Journal of Ecology 98: 1168–1177. https://doi.org/10.1111/j.1365-2745.2010.01683.x.
Total of 108 references.

