Influence of natural habitat fragmentation on the genetic structure of Canarian populations of Juniperus turbinata
Jimenez J. F., Sánchez-Gómez P., Cánovas J. L., Hensen I., Aouissat M. (2017). Influence of natural habitat fragmentation on the genetic structure of Canarian populations of Juniperus turbinata. Silva Fennica vol. 51 no. 2 article id 1678. https://doi.org/10.14214/sf.1678
Highlights
- Canarian populations of Juniperus turbinata exhibited levels of genetic diversity similar to those of mainland populations
- Despite the historical geologic events occurred in Canary Islands, and the documented decrease of populations since the arrival of settlers, no genetic differentiation between islands has been observed
- As it has been observed for other Macaronesian endemics, J. turbinata long dispersal ability seems to be the main force driving the genetic structure of populations.
Abstract
Oceanic archipelagos provide an important platform from which to evaluate the effects of isolation and fragmentation on the genetic structure of species. As a result of oceanic isolation, such species usually show lower levels of genetic diversity and higher genetic differentiation than their mainland congeners. However, this is not necessarily the case for long distance dispersal species, whose genetic structure is not strictly defined by population isolation. We assessed the level and distribution of genetic diversity among Canarian populations of Juniperus turbinata in order to evaluate the influence of population isolation on its genetic structure. Using Amplified Fragment Length Polymorphism markers, we analyzed molecular diversity among 175 individuals from five populations occurring across the Canary Island and three Moroccan populations. Principal Coordinate Analysis, neighbor joining clustering, AMOVA and Bayesian-based analysis were applied to examine population structure. Despite the documented habitat loss and decline in Canarian populations, Amplified Fragment Length Polymorphism markers revealed levels of intra-population genetic diversity that were similar to those from mainland populations, and low levels of genetic differentiation. Bayesian analysis of population structure showed three main clusters, one comprising El Hierro population and a few individuals from several islands, a second cluster that grouped the remaining Canarian populations together, and a third cluster grouping Moroccan populations. Our results suggest that the main force driving the genetic structure of Canarian populations of J. turbinata is its capacity for long distance dispersal.
Keywords
genetic diversity;
AFLP markers;
Canarian juniper;
oceanic islands
-
Jimenez,
Departamento de Biología Vegetal (Botánica), Universidad de Murcia, Campus de Espinardo s/n, E-30100 Murcia, Spain
E-mail
fjimenez@um.es

-
Sánchez-Gómez,
Departamento de Biología Vegetal (Botánica), Universidad de Murcia, Campus de Espinardo s/n, E-30100 Murcia, Spain
 http://orcid.org/0000-0001-6754-1512
E-mail
psgomez@um.es
http://orcid.org/0000-0001-6754-1512
E-mail
psgomez@um.es
- Cánovas, Departamento de Biología Vegetal (Botánica), Universidad de Murcia, Campus de Espinardo s/n, E-30100 Murcia, Spain E-mail joseluis.canovas@um.es
- Hensen, Institut für Biologie, Martin-Luther Universitat, 06099 Halle, Germany E-mail isabell.hensen@botanik.uni-halle.de
- Aouissat, Centre Universitaire Salhi Ahmed Naama, BP 66, Naâma, Algérie E-mail aouissatm@yahoo.fr
Received 12 July 2016 Accepted 2 March 2017 Published 9 March 2017
Views 159874
Available at https://doi.org/10.14214/sf.1678 | Download PDF

1 Introduction
Juniperus phoenicea group (family Cupressaceae) is an assortment of small monoecious or rarely dioecious Mediterranean trees whose distribution extends encompasses the Mediterranean basin and Macaronesian regions, from Madeira and Canary Islands in the west to Jordan and Saudi Arabia in the east, and is most prevalent throughout the Iberian Peninsula and North Africa (Quézel and Pesson 1980; Greuter et al. 1984; do Amaral Franco 1986; Farjon 2005). The species comprises two subspecies, J. phoenicea subsp. phoenicea and J. phoenicea subsp. turbinata (Guss.) Nyman, which were defined based on taxonomic (Farjon 2005), morphological (Mazur et al. 2010) and molecular studies (Adams et al. 2002; Boratyński et al. 2009; Dzialuk et al. 2011). Alternative interpretations define two independent species, J. phoenicea s. str. and J. turbinata Guss., with J. turbinata being discussed by several authors (see Farjon 2005), and its recognition as a separate being supported by morphological and biochemical studies (Lebreton and Pérez de Paz 2001; Mazur et al. 2016) and recent molecular studies (Adams and Schwarzbach 2013; Adams 2014; Adams et al. 2013, 2014). As such, J. phoenicea ≡ J. phoenicea subsp. phoenicea is generally considered as being endemic to the Iberian Peninsula, Northwest Italy and the south of France, while J. turbinata ≡ J. phoenicea subsp. turbinata is widespread throughout the Mediterranean basin and Macaronesia. Furthermore, while the Macaronesian populations of J. turbinata were formerly described as J. canariensis Guyot & Mathou, they were later combined as a subspecies, J. turbinata subsp. canariensis (Guyot & Mathou) Rivas Mart., Wildpret & P. Pérez by Rivas-Martínez et al. (1993). However, its taxonomic status has recently been challenged based on several molecular markers (RAPDs, Adams et al. 2006; cpDNA, Adams et al. 2010; nrDNA and cpDNA, Adams et al. 2013a), leading to the conclusion that J. turbinata subsp. canariensis should not be supported at the subspecific level.
Regardless of whether Macaronesian populations represent a well-defined subspecies, they are scarce and threatened, and the islands where they occur are largely considered priority natural habitats by the European Directive 92/43, due to the particular richness of endemic species (Otto et al. 2012). At present, Canarian populations of J. turbinata are located in the five westernmost islands of the archipelago, and only occupy some 10% of the species potential distribution range due to widespread human disturbance (Fernández-Palacios et al. 2008; Del Arco et al. 2010). The taxon is reported as having been formerly widespread throughout the Canary Islands (including the eastern islands of Lanzarote and Fuerteventura), until their colonization, approximately six centuries ago. Populations of Tenerife, La Palma and Gran Canaria are usually small and frequently formed by isolated individuals, whereas the largest populations are located on La Gomera and El Hierro islands. Del Arco et al. (2010) calculated a distribution area of 3943 ha for J. turbinata across the Canary Islands, with 70% occurring in La Gomera.
Although there are several studies that point out the dynamics of Canarian populations of J. turbinata (Fernández-Palacios et al. 2008; Otto et al. 2012), there is a lack of knowledge on the genetic structure of these populations. As such, research into the amount and distribution of genetic diversity is required to a better understanding of associated ecological processes such as island colonization, gene flow among populations or historical bottlenecks, and to enhance and inform existing conservation measures aimed at protecting the populations (Fernández-Palacios et al 2008; Otto et al. 2010).
It is well known that oceanic islands provide ample opportunity to test hypotheses on the ecological factors affecting species’ genetic structure. These may include colonization patterns, species origin/diversity, island biogeography and population differentiation (Francisco-Ortega et al. 2000; Emerson 2002; Stuessy et al. 2014). On oceanic islands, taxa are usually somewhat differentiated from those of mainland populations and they tend to display lower levels of genetic diversity because each island can be considered as an isolated subpopulation, and the potential area of population distribution is restricted, so population sizes are usually limited, making populations more prone to genetic risks, such as genetic drift, loss of genetic diversity or even extinction by stochastic factors or human disturbance (Ellstrand and Elam 1993; Franckham 1997; Whittaker et al. 2008). In addition, genetic differentiation is higher between oceanic islands than within them and populations mostly exhibit isolation-by-distance patterns of differentiation (Franckham 1997; Whittaker and González-Palacios 2007). However, some studies have also shown similar or even higher levels of genetic variation in widespread island taxa in comparison with mainland relatives (Francisco-Ortega et al. 2000; Su et al. 2010). In species capable of long-distance dispersal genetic differentiation among islands may be reduced, and intra-population genetic diversity might be higher than otherwise expected (García Verdugo et al. 2014; García-Verdugo et al. 2015).
With a view to contributing to more global study on J. phoenicea, for the current study, we report on J. turbinata using AFLP markers. Our main aim was to determine the genetic structure of the Canarian population revealed by neutral nuclear markers, and to evaluate whether its structure is more generally affected by geographical isolation or by gene flow. As such, subpopulations of J. turbinata were analyzed: a) to assess genetic diversity across the Canarian populations, and b) to infer the influence of isolation in the genetic structure of J. turbinata.
2 Material and methods
2.1 Plant Sampling
As J. turbinata occurs mainly as dispersed individuals in the Canary Islands, the strategy was to sample at locations in which individuals were grouped forming populations. So, a population from each of the five Canarian islands where J. turbinata occurs was sampled. Three populations from mainland Morocco were also sampled to compare values of genetic diversity with those obtained for the island populations. So, 175 individuals belonging to eight populations were sampled. The locations of sampled populations are shown in Fig. 1 and their characteristics are listed in Table 1. To avoid damaging individuals, the sampling strategy consisted of collecting apexes of branches measuring about 10 cm in length. Furthermore, we only sampled mature individuals that were sufficiently spaced. All plant material was stored in labeled ziplock bags with silica gel at room temperature to dry it until DNA extraction (Chase and Hills 1991).
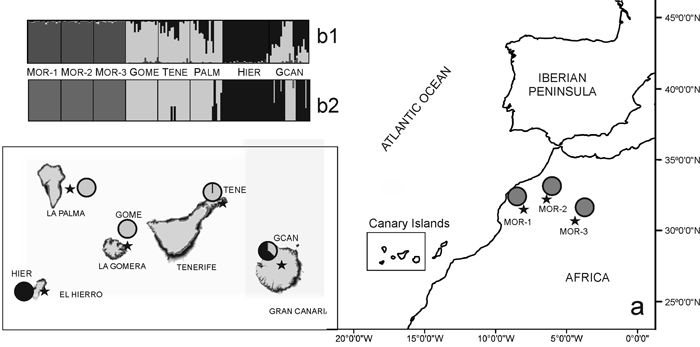
Fig. 1. (a) Location map and Bayesian analysis of the genetic structure of 175 individuals from eight sampled populations of Juniperus turbinata. The great rectangle that includes Canary Islands represent an enlargement of the same area in the small rectangle Pie charts represent the mean proportion of membership of each population to the predefined K = 3 clusters with BAPS. (b) Plots show the membership of individuals to the predefined K = 3 clusters obtanined with b1, STRUCTURE and b2, BAPS. Number and codes of populations are listed in Table 1. View larger in new window/tab.
| Table 1. Geographical location of eight sampled populations of Juniperus turbinata and genetic diversity estimates. N = Number of individuals sampled per population; PLP = Percentage of polymorphic loci; He = Expected heterozygosity (Nei’s gene diversity); Hs = Mean Nei’s gene diversity within populations; Ht = Total gene diversity; Fst = Index of population differentiation. | ||||||||
| Pop | Island/*Country | Location | N | PLP | He | Hs | Ht | Fst |
| MOR-1 | Morocco* | 31°35´N, 07°76´W | 20 | 67.7 | 0.217 | |||
| MOR-2 | Morocco* | 32°18´N, 06°53´W | 20 | 65.0 | 0.207 | |||
| MOR-3 | Morocco* | 32°61´N, 04°53´W | 20 | 66.2 | 0.203 | |||
| GOME | La Gomera | 28°19´N, 17°26´W | 20 | 69.6 | 0.214 | |||
| TENE | Tenerife | 28°56´N, 16°24´W | 20 | 68.3 | 0.213 | |||
| PALM | La Palma | 28°68´N, 17°77´W | 20 | 73.7 | 0.218 | |||
| HIER | El Hierro | 27°75´N, 18°13´W | 30 | 68.4 | 0.192 | |||
| GCAN | Gran Canaria | 28°01´N, 15°65´W | 25 | 67.4 | 0.211 | |||
| TOTAL | 175 | 94.1 | 0.209 | 0.243 | 0.1398 | |||
2.2 DNA analyses
DNA was extracted from silica gel dried leaves using the 2 × cetyltrimethylammonium bromide (CTAB) method (Doyle and Doyle 1987). Total DNA extracts were quantified using a Nanodrop 2000 spectrophotometer. Amplified fragment length polymorphism (AFLP) analysis was carried out following Vos et al. (1995) with minor modifications as follows. The analysis was performed with fluorescence-labeled primers (VIC, NED, PET, Applied Biosystems, Madrid, Spain) instead of radioactive labeled primers. Fragments were selectively amplified with the primer pairs EcoR1-ATG/MseI-CGT, EcoRI-ACA/MseI-CGT, EcoRI-ACG/MseI-CAC. Multiplex products were run for 4 h on an ABI 377 sequencer to separate fragments together with an internal size standard (GeneScan 600 LIZ, ABI). Sizing and peak identification were performed using Genemapper 4.0 software (Applied Biosystems). In the finally assembled binary matrix the presence of a band was scored as 1 while the absence of a band was scored as 0.
As AFLP markers are dominant, we assumed that null bands were homologous and that populations were in Hardy-Weinberg equilibrium (Lynch and Milligan 1994) in order to compute diversity indices [percentage of polymorphic markers (PLP) and Nei’s (1978) unbiased expected heterozygosity (He)] and genetic distance among populations (Fst). These parameters were inferred with AFLP-surv 1.0 (Vekemans 2002). Significance of Fst values was determined using 1000 bootstrapped data sets.
A Bayesian model-based analysis was performed to infer population structure with Structure version 2.2 (Pritchard et al. 2000; Falush et al. 2007). The F model, based on an admixture ancestry model with correlated allele frequencies, was imposed to estimate the posterior probabilities [LnP(D)] of K groups (Pritchard and Wen 2004) and the individual percentages of membership assigned to them according to their molecular multilocus profiles (Falush et al. 2003, 2007). Probabilities for a range of K were examined starting from one to the number of sampled populations plus one (K = 1–9), using a burn-in period and run length of the Markov chain Monte Carlo (MCMC) of 105 and 106 iterations, respectively, replicated 20 times. Results were uploaded into STRUCTURE HARVESTER (Earl and von Holdt 2012, available at http://taylor0.biology.ucla.edu/struct_harvest/), which estimates the most likely K value (ΔK), following Evanno et al. (2005). We used CLUMPP 1.1.2 (Jakobsson and Rosenberg 2007) to reach a consensus on the results of the independent runs for the optimal K. For the consensus, we used the Greedy option with random input order and 10 000 repeats. The consensus was visualized in DISTRUCT 1.1 (Rosenberg 2004). Alternatively, another bayesian analysis of the genetic structure was conducted with BAPS 5.4 (Corander and Marttinen 2006), using stochastic optimization instead of Markov chain Monte Carlo to find the optimal partition. We performed a mixture analysis of individuals without using the geographic origin of the samples as a prior informative (clustering of individuals). BAPS simulations were run with the maximal number of groups (K) set from 1 to 10. Each run was replicated 10 times, and results were averaged according to the resultant likelihood scores. The output of the mixture analysis was used as the input for population admixture analysis (Corander and Marttinen 2006), with the default settings to detect admixture between clusters.
In addition, two Principal Coordinate Analyses (PCoA) were performed to illustrate overall similarity among individuals using Multivariate Statistical Package software (MVSP version 3.12d http://www.kovcomp.com/mvsp). The first one included all the sampled individuals, while the second one was performed using only Canarian populations, in order to detect any substructure of genetic diversity. PCoA was inferred from the pairwise Nei’s genetic distance (Nei 1978) between all pairs of AFLP phenotypes.
Pairwise Fst-values between all populations were also calculated and tested for significance by resampling with 1000 random permutations. Mantel tests (Mantel 1967) were carried out to assess correlation among genetic and geographic distances. These analyses were performed using all populations, and alternatively using only Canarian populations. Thereafter, the fine-scale genetic structure among the Canarian populations was investigated using spatial autocorrelation analysis in GenAlEx 6.5 (Peakall and Smouse 2006). The spatial autocorrelation coefficient (r) was computed using the geographic distance and the genetic distance between individuals. Tests for statistical significance were conducted by random shuffling (999 times) of individual geographic locations to define the upper and lower bounds of the 95% confidence interval for each distance class, and estimating 95% confidence intervals around mean r values by bootstrapping pair-wise comparisons within each distance class (1000 repeats). Analyses were performed using the variable distance classes option based on the geographical distances estimated in each population.
3 Results
The three selective AFLP primer combinations amplified a total of 589 fragments. In total 94.1% of the fragments were polymorphic and every of 175 individuals had unique AFLP profiles. No private markers for island were detected, but we found five exclusive bands for Canary Islands, which appeared in more than 80% of sampled individuals, and two exclusive bands for the Moroccan populations. The proportion of polymorphic loci (PLP) for a single population ranged from 65.0% (MOR2) to 73.7% (PALM). Expected heterozygosity values showed that PALM was the most diverse population (He = 0.218), whereas HIER showed the lowest within-population diversity (0.192). The average gene diversity within populations (Hs) was 0.209, whereas the total diversity (Ht) value was 0.243 (Table 1). The fixation index was highly significant (Fst = 0.1398, P < 0.000), suggesting low to moderate genetic differentiation among populations.
The Bayesian analysis of genetic structure of J. turbinata populations conducted with Structure found the highest LnP(D) and ΔK values for K = 3. Moroccan populations were assigned to one cluster and Canarian populations were assigned to other two clusters; the first of them grouped individuals of GOME, TENE, and PALM with high proportion of membership and the last cluster grouped HIER, whereas GCAN individuals showed proportion of membership intermediate between these two clusters. The genetic structure revealed by BAPS was highly congruent with STRUCTURE results identifying three clusters (P = 0.999). However, only 5.14% of individuals were admixed (Fig. 1). The first cluster grouped the individuals from Morocco. The second one grouped GOME, TENE, PALM and seven individuals from GCAN, whereas the third cluster included HIER and 18 individuals from GCAN. The individuals admixed were four from GCAN, three from PALM and two from TENE. PCoA analyses gave similar results (Fig. 2). PCoA plot performed including Moroccan population revealed two clusters, one comprising the samples of Morocco and a second cluster that grouped Canarian populations (Fig. 2a). For this analysis the three first axes accounted for 22.76% of the variation (15.51%, 4.33% and 2.92%, respectively). PCoA analysis for the Canarian populations revealed three clusters (three first axes explained 8.67%, 4.05% and 2.80% of the variation respectively). The first cluster was constituted by individuals from TENE, GOME and GCAN. The second grouped individuals form populations of TENE, GOME, GCAN and PALM. The third was formed by HIER population, including some individuals from GCAN (Fig. 2b).
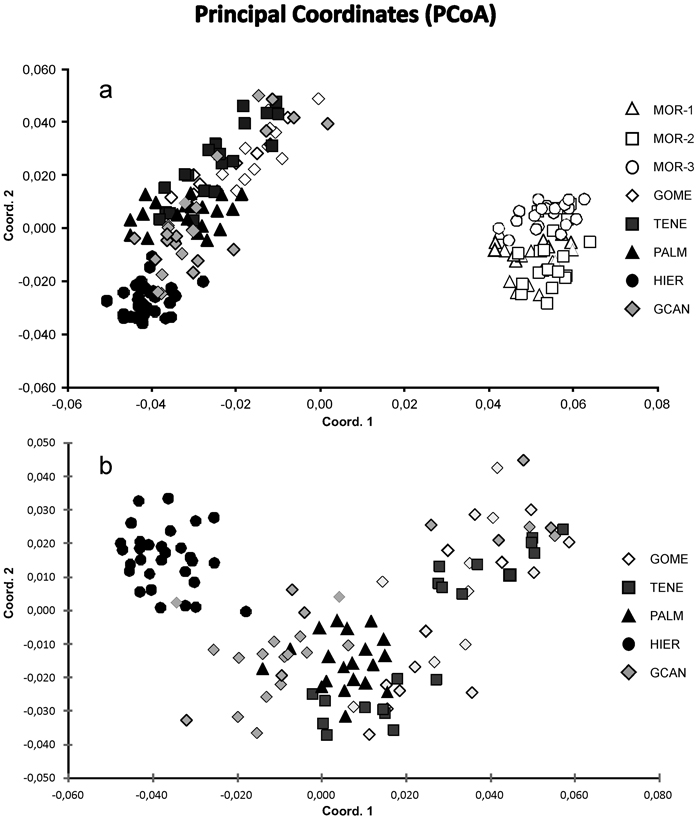
Fig. 2. Principal coordinate analysis of the 175 individuals from eight sampled populations of J. turbinata based on pair-wise Nei’s (1978) genetic distances. (a) Scatter plot showing eight populations based on two first components of the PCoA. (b) Scatter plot showing 115 individuals from five Canarian populations. Number and codes of populations are listed in Table 1.
Pairwise Fst among populations showed high differentiation between Moroccan and Canarian populations. In addition HIER population was moderately differentiated from TENE and GOME populations (Table 2). The remaining populations revealed low pairwise genetic differentiation. The Mantel test in which were included Moroccan populations showed a great isolation by distance pattern (R = 0.909, P = 0.001). When these populations were eliminated from the analysis the correlation among genetic and geographic distances was not significant (R = 0.301, P = 0.085). Spatial autocorrelation analysis showed positive isolation by distance at distances of 90 km approximately (Fig. 3).
| Table 2. Pairwise estimate Fst values among eight populations of J. turbinata. All values are significant at 0.001 level. Number and codes of populations are listed in Table 1. | ||||||||
| MOR-1 | MOR-2 | MOR-3 | GOME | TENE | PALM | HIER | GCAN | |
| MOR-1 | 0.000 | |||||||
| MOR-2 | 0.042 | 0.000 | ||||||
| MOR-3 | 0.067 | 0.051 | 0.000 | |||||
| GOME | 0.152 | 0.159 | 0.155 | 0.000 | ||||
| TENE | 0.185 | 0.192 | 0.197 | 0.016 | 0.000 | |||
| PALM | 0.193 | 0.206 | 0.212 | 0.037 | 0.046 | 0.000 | ||
| HIER | 0.254 | 0.265 | 0.276 | 0.120 | 0.114 | 0.071 | 0.000 | |
| GCAN | 0.181 | 0.190 | 0.188 | 0.033 | 0.039 | 0.021 | 0.068 | 0.000 |
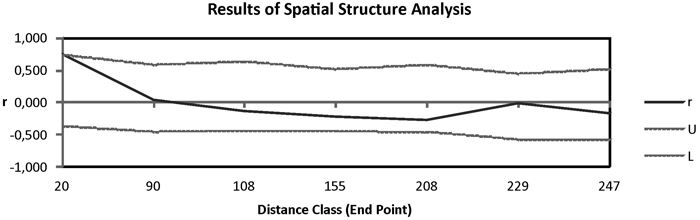
Fig. 3. Correlogram from spatial autocorrelation analysis using the correlation coefficient r, and variable distance classes. 95% confidence error bars for r were estimated by bootstrapping over pairs of samples; dashed lines (U, L) represent upper and lower bounds of a 95% CI for r generated under the null hypothesis of random geographic distribution.
4 Discussion
4.1 Genetic diversity
It is well documented that the genetic diversity of small island species is usually lower than those of their mainland congeners (Frankham 1997; Whittaker et al. 2008), which is a result of founder effects or bottlenecks. In our study, genetic diversity estimates obtained with AFLP do not follow predicted patterns. Although Canarian populations are differentiated from Moroccan populations, PLP and He suggest that genetic diversity is not impoverished. This evidence is not unusual, as García-Verdugo et al. (2015) suggest that in more than 50% of studies in which genetic diversity of island plants is addressed, island populations show similar or higher genetic diversity than their mainland relatives, with possible causes being the ability of some plant species to recurrently overcome sea barriers, or the role of islands as climatic refugia (García-Vergudo et al. 2015 and references therein). With respect to other studies on the genetic diversity of J. turbinata populations is acknowledged that genetic diversity values obtained with molecular markers are difficult to compare between studies, because the statistical methods used to process the data often vary, particularly with dominant and co-dominant markers. However, parameters such as intra-population genetic markers may serve to establish comparisons between species (Nybom 2004). Regarding the He values found in our study, studies on J. phoenicea group by Boratyński et al. (2009) and Dzialuk et al. (2011) produced similar results to ours, while Meloni et al. (2006) obtained higher Hs values. The intra-population genetic diversity observed in our study falls within the range of the values reported by Nybom (2004) for outcrossing, long-lived endemic species.
Our results therefore imply that, despite the low number of individuals sampled, as well as the documented historical decrease of populations throughout the Canarian archipelago (Fernández-Palacios et al. 2008), J. turbinata has moderate levels of genetic diversity (evidenced by AFLP) with no apparent evidence pointing to genetic bottlenecks. Long-lived woody perennials are expected to be resilient to changes in genetic diversity due to ample gene flow and long generation times. Furthermore, fleshy fruited species typically show widespread distributions coupled with extensive gene flow among islands (García-Verdugo et al. 2014). Therefore, Canarian populations of J. turbinata may have been able to avoid the effects of genetic drift through long-distance dispersal, which has previously been described for several Macaronesian species (Ferreira et al. 2001; Vargas 2007; García-Verdugo et al. 2010; García-Verdugo et al. 2014; Rumeu et al. 2014).
4.2 The genetic structure of populations
The Fst pairwise estimates obtained in this study showed high levels of genetic differentiation between MOR and Canarian populations and low to moderate population differentiation among Canarian populations (paiwise Fst < 0.13). Furthermore, the Mantel test and spatial autocorrelation analysis failed to predict an isolation-by-distance pattern when only Canarian populations were analyzed. This pattern of differentiation suggests an ancient colonization of the Canary Islands by mainland populations, with a posterior interruption of gene flow with African populations. This is plausible if we taking into account that the estimated age of J. phoenicea s.l. is more than 30 Mya (Mao et al. 2010). According to the order of emergence of islands, it is reasonable to think that colonization occurred from eastern to western islands. However, with our available data is difficult predict if it was a single or multiple colonization events, or the role of each island in these hypothetical events. The levels of genetic differentiation between mainland and island populations suggest an anagenetic type of evolution for the Canarian populations, in which genetic variation would have been accumulating for generations through mutation and recombination until the overall level of genetic variation may approximate to that of source population (Stuessy et al. 2006).
However, the distribution of genetic variation suggests substantial gene flow between almost all the islands, which seems to be less effective between El Hierro and the others. Surprisingly, the PCoA, the admixed individuals depicted by BAPS and the pairwise Fst suggest that the Gran Canaria population is most similar to the El Hierro population, which is the most geographically remote of the islands. This finding would suggest more complex patterns of gene flow among islands have taken place than the predicted stepping-stone model of differentiation from east to west, such as that observed for Gallotia spp. (González et al. 1996) or Olea europaea L. (Hess et al. 2000). Historical geologic events might have played an important role in the partitioning of genetic diversity by promoting local extinctions and re-colonizations, as it has been observed for several groups of animals and plants (Brown and Pestano 1998; Nogales et al. 1998; Juan et al. 2000; Fernández-Mazuecos and Vargas 2011). Unfortunately, due to the scarcity of well defined populations in Canary Islands, we sampled a population per island, instead individuals from different locations in each island. Therefore, observed genetic differences could be due to this sampling strategy. Adding georeferenced individuals from different locations in further studies might elucidate patterns of gene flow among islands, colonization events, etc.
Furthermore, the documented arrival of settlers to Canary Islands might have been a cause of intra-population genetic diversity decrease and differentiation increase, through the transformation of favourable habitats in grazing (Fernández Palacios et al. 2011). However, as demonstrated for several Canarian endemics (García-Verdugo et al. 2010; Ferreira et al. 2011; Rumeu et al. 2011; Sosa et al. 2013), the recurrent gene flow between islands have revealed moderate levels of genetic diversity and absence of genetic structure across islands. The ability of J. phoenicea s.l. to disperse both pollen (by wind) and seeds (by birds) to distant locations has already been documented in mainland populations (Jordano 1993). The known dispersers of J. turbinata in Canary Islands are ravens, blackbirds and lizards (Nogales et al. 1999; Fernández-Palacios et al. 2008; Otto et al. 2010). Long distance dispersal (LDD) mechanisms in oceanic islands involves several types of dispersal; anemochory, zoochory, by birds capable to fly over long distances, as well as diplochory or secondary seed dispersal, in which the primary disperser, is hunted by a predator capable to travel between islands, and sea dispersal or hydrochory. (Nogales et al. 2012; Vargas et al. 2014). In this case dispersal by birds and diplochory might be effective methods of LDD (Nogales et al. 1999, 2002, 2007; Padilla et al. 2012). Furthermore, although J. turbinata do not exhibit specialized floating structures, dry galbulus with fertile seeds are capable to float, so hydrochory could also be considered as an effective LDD mechanism. These abilities could have led to an important interexchange of pollen and seeds between islands that may mask any ancient relationships.
As yet, while we do not have any data on the distribution of cpDNA haplotypes for our study populations, such information may enable us to infer the underlying processes driving colonization more accurately, as previously demonstrated for several other Canarian endemics such as J. cedrus Webb. & Berth. (Rumeu et al. 2014), or O. europaea subsp. guanchica P. Vargas, J. Hess, Muñoz Garm. & Kadereit (García Verdugo et al. 2009). Further studies are therefore recommended to shed more light on the specific processes underlying island colonization.
Adknowledgements
Authors wish to thank to David López García and Juan Bautista Vera for field assistance in collecting samples. This work has been supported by a Ministry of Economy and Competitiveness CGL2011-30099 project grant to Pedro Sánchez Gómez.
References
Adams R.P. (2014). Junipers of the world: the genus Juniperus, 4th ed. Trafford Publ. Bloomington, IN.
Adams R.P., Schwarzbach A.E. (2013). Phylogeny of Juniperus using nrDNA and four cpDNA regions. Phytologia 95: 179–187.
Adams R.P., Padney N., Rezzi S., Casanova J. (2002). Geographic variation in the random amplified polymorphic DNAs (RAPDs) of Juniperus phoenicea, J. p. var. canariensis, J. p. subsp. eumediterranea and J. p. var turbinata. Biochemichal Systematics and Ecology 30(3): 223–229. https://doi.org/10.1016/S0305-1978(01)00083-7.
Adams R.P., Nguyen S., Achak N. (2006). Geographic variation in Juniperus phoenicea (Cupressaceae) from the Canary Islands, Morocco and Spain based on RAPDs analysis. Phytologia 88: 270–278.
Adams R.P., Rumeu B.R., Nogales M., Fontinha S.S. (2010). Geographic variation and systematics of Juniperus phoenicea L. from Madeira and the Canary Islands: analyses of SNPs from nrDNA and petN-psbM DNA. Phytologia 92: 59–67.
Adams R.P., Boratyński A., Arista M., Schwarzbach A.E., Leschner H., Liber Z., Minissale P., Mataraci T., Manolis A. (2013). Analysis of Juniperus phoenicea from throughout its range in the Mediterranean using DNA sequence data from nrDNA and petN-psbM: the case for the recognition of J. turbinata Guss. Phytologia 95: 202–209.
Adams R.P., Altarejos J., Arista M., Schwarzbach A.E. (2014). Geographic variation in J. phoenicea var. phoenicea from throughout its range: analysis of nrDNA and the petN-PsbM cp region. Phytologia 96: 247–251.
Boratyński A., Lewandowski A., Boratyńska K., Montserrat J.M., Romo A. (2009). High level of genetic differentiation of Juniperus phoenicea (Cupressaceae) in the Mediterranean region: geographic implications. Plant Systematics and Evolution 277(3): 163–172. https://doi.org/10.1007/s00606-008-0122-z.
Brown R.P., Pestano J. (1998) Phylogeography of skinks (Chalcides) in the Canary Islands inferred from mitochondrial DNA sequences. Molecular Ecology 7(9): 1183–1191. https://doi.org/10.1046/j.1365-294x.1998.00442.x.
Chase M.W., Hills H.H. (1991). Silica gel: an ideal material for field preservation of leaf samples for DNA studies. Taxon 40(2): 215–220. https://doi.org/10.2307/1222975.
Corander J., Marttinen P. (2006). Bayesian identification of admixture events using multi-locus molecular markers. Molecular Ecology 15(10): 2833–2843. https://doi.org/10.1111/j.1365-294X.2006.02994.x.
Del Arco M., González-González R., Garzón-Machado V., Pizarro-Hernández B. (2010). Actual and potential natural vegetation on the Canary Islands and its conservation status. Biodiversity and Conservation 19(11): 3089–3140. https://doi.org/10.1007/s10531-010-9881-2.
do Amaral Franco J. (1986). Juniperus L. In: Castroviejo S., Laínz M., López González G., Montserrat P., Muñoz Garmendia F., Paiva J., Villar L. (eds.). Flora iberica, vol 1. Real Jardín Botánico, C.S.I.C. Madrid. Spain. p. 181–188.
Doyle J.J., Doyle J.L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochemical Bulletin 19: 11–15.
Dzialuk A., Mazur M., Boratyńska K., Monstserrat J.M., Romo A., Boratyński A. (2011). Population genetic structure of Juniperus phoenicea (Cupressaceae) in the western Mediterranean Basin: gradient of diversity on a broad geographical scale. Annals of Forest Science 68(8): 1341–1350. https://doi.org/10.1007/s13595-011-0150-7.
Earl D.A., von Holdt B.M. (2012). Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4(2): 359–361. https://doi.org/10.1007/s12686-011-9548-7.
Ellstrand N.C., Elam D.R. (1993). Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics 24: 217–242. https://doi.org/10.1146/annurev.es.24.110193.001245.
Emerson B.C. (2002). Evolution on oceanic islands: molecular phylogenetic approaches to understanding pattern and process. Molecular Ecology 11(6): 951–966. https://doi.org/10.1046/j.1365-294X.2002.01507.x.
Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software structure: a simulation study. Molecular Ecology 14(8): 2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x.
Falush D., Stephens M., Pritchard J.K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Falush D., Stephens M., Pritchard J.K. (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes 7(4): 574–578. https://doi.org/10.1111/j.1471-8286.2007.01758.x.
Farjon A. (2005). A monograph of Cupressaceae and Sciadopitys. Royal Botanic Gardens, Kew, London. 648 p.
Fernández-Mazuecos M., Vargas P. (2011). Genetically depauperate in the continent but rich in oceanic islands: Cistus monspeliensis (Cistaceae) in the Canary Islands. PLoS ONE 6(2): e17172. https://doi.org/10.1371/journal.pone.0017172.
Fernández Palacios J.M., Otto R., Delgado J.D., Arévalo J.R., Naranjo A., González Artiles F., Morici C., Barone R. (eds.) (2008). Los Bosques Termófilos de Canarias. Proyecto LIFE04/NAT/ES/000064. [Canarian termophilic forests. Project. LIFE04/NAT/ES/000064]. Excmo. Cabildo Insular de Tenerife, Santa Cruz de Tenerife, Spain. 202 p.
Fernández Palacios J.M., de Nascimento L., Otto R., Delgado J.D., García-del-Rey E., Arévalo J.R., Whittaker R.J. (2011). A reconstruction of Palaeo-Macaronesia, with particular reference to the long-term biogeography of the Atlantic island laurel forests. Journal of Biogeography 38(2): 226–246. https://doi.org/10.1111/j.1365-2699.2010.02427.x.
Ferreira R.C., Piredda R., Bagnoli F., Bellarosa R., Attimonelli M., Fineschi S., Schirone B., Simeone M.C. (2011). Phylogeography and conservation perspectives of an endangered Macaronesian endemic: Picconia azorica (Tutin) Knobl. (Oleaceae). European Journal of Forest Research 130(2): 181–195. https://doi.org/10.1007/s10342-010-0420-1.
Francisco-Ortega J., Santos-Guerra A., Kim S.-C., Crawford D.J. (2000). Plant genetic diversity in the Canary Islands: a conservation perspective. American Journal of Botany 87(7): 909–919. https://doi.org/10.2307/2656988.
Frankham R. (1997). Do island populations have less genetic variation than mainland populations? Heredity 78: 311–327. https://doi.org/10.1038/hdy.1997.46.
García-Verdugo C., Fay M.F., Granado-Yela C., Rubio de Casas R.R., Balaguer L., Besnard G., Vargas P. (2009). Genetic diversity and differentiation processes in the ploidy series of Olea europaea L.: a multiscale approach from subspecies to insular populations. Molecular Ecology 18(3): 454–467. https://doi.org/10.1111/j.1365-294X.2008.04027.x.
García-Verdugo C., Forrest A.D., Fay M.F., Vargas P. (2010). The relevance of gene flow in metapopulation dynamics of an oceanic island endemic, Olea europaea subsp. guanchica. Evolution 64(12): 3525–3536. https://doi.org/10.1111/j.1558-5646.2010.01091.x.
García-Verdugo C., Baldwin B.G., Fay M.F., Caujapé-Castell J. (2014). Life history traits and patterns of diversification in oceanic archipelagos: a meta-analysis. Botanical Journal of the Linnean Society 174(3): 334–348. https://doi.org/10.1111/boj.12127.
García-Verdugo C., Sajeva M., La Mantia T., Harrouni C., Msanda F., Cajuapé-Castell J. (2015). Do island plant populations really have lower genetic variation than mainland populations? Effects of selection and distribution range on genetic diversity estimates. Molecular Ecology 24(4): 726–741. https://doi.org/10.1111/mec.13060.
González P., Pinto F., Nogales M., Jiménez-Asensio J., Hernández M., Cabrera V.M. (1996). Phylogenetic relationships of the Canary Islands endemic lizard Gallotia (Sauria: Lacertidae), inferred from mitochondrial DNA sequences. Molecular and Phylogenetic Evolution 6(1): 63–71. https://doi.org/10.1006/mpev.1996.0058.
Greuter W., Burdet H.M., Long G. (1984). Med.-Checklist 1. Conservatoire et Jardin Botaniques de la Ville de Genève et Med.-Checklist Trust of OPTIMA, Genève.
Hess J., Kadereit J.W., Vargas P. (2000). The colonization history of Olea europaea L. in Macaronesia based on internal transcribed spacer 1 (ITS-1) sequences, randomly amplified polymorphic DNAs (RAPD), and intersimple sequence repeats (ISSR). Molecular Ecology 9(7): 857–868. https://doi.org/10.1046/j.1365-294x.2000.00942.x.
Jakobsson M., Rosenberg N.A. (2007). CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23(14): 1801–1806. https://doi.org/10.1093/bioinformatics/btm233.
Jordano P. (1993). Geographic ecology and variation of plant-seed disperser interaction: southern Spanish junipers and frugivorous thrushes. Vegetatio 107/108: 85–104.
Juan C., Emerson B.C., Oromí P., Hewitt G.M. (2000). Colonization and diversification: towards a phylogeographic synthesis for the Canary Islands. Trends in Ecology & Evolultion 15(3=: 104–109. https://doi.org/10.1016/S0169-5347(99)01776-0.
Lebreton P., Pérez de Paz P.L. (2001). Définition du Genévrier de Phénicie (Juniperus aggr. phoenicea), reconsidéré à ses limites biogéographiques: Méditerranée orientale (Crète et Chypre) et Atlantique (Iles Canaries). Bulletin mensuel de la Société linnéenne de Lyon 70(4): 73–92. https://doi.org/10.3406/linly.2001.11373.
Lynch M., Milligan B.G. (1994). Analysis of population genetic structure with RAPD markers. Molecular Ecology 3(2): 91–99. https://doi.org/10.1111/j.1365-294X.1994.tb00109.x.
Mantel N. (1967). The detection of disease clustering and a generalized regression approach. Cancer Research 27: 209–220.
Mao K., Hao G., Liu J., Adams R.P., Milne R.I. (2010). Diversification and biogeography of Juniperus (Cupressaceae): variable diversification rates and multiple intercontinental dispersals. New Phytologist 188(1): 254–272. https://doi.org/10.1111/j.1469-8137.2010.03351.x.
Mazur M., Klajbor K., Kielich M., Sowińska M., Romo A., Montserrat J.M., Boratyński A. (2010). Intra-specific differentiation of Juniperus phoenicea in the western Mediterranean region revealed in morphological multivariate analysis. Dendrobiology 63: 21–31.
Mazur M., Minissale P., Sciandrello S., Boratyński A. (2016). Morphological and ecological comparison of populations of Juniperus turbinata Guss. and J. phoenicea L. from the Mediterranean region. Plant Biosystems 150(2): 313–322. https://doi.org/10.1080/11263504.2014.994579.
Meloni M., Perini D., Filigheddu R., Binelli G. (2006). Genetic variation in five Mediterranean populations of Juniperus phoenicea as revealed by inter-simple sequence repeat (ISSR) markers. Annals of Botany 97(2): 299–304. https://doi.org/10.1093/aob/mcj024.
Nei M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590.
Nogales M., López M., Jiménez-Asensio J., Larruga J.M., Hernández M., González P. (1998). Evolution and biogeography of the genus Tarentola (Sauria: Gekkonidae) in the Canary Islands, inferred from mitochondrial DNA sequences. Journal of Evolutionary Biology 11(4): 481–494. https://doi.org/10.1046/j.1420-9101.1998.11040481.x.
Nogales M., Hernández E.C., Valdés F. (1999). Seed dispersal by common ravens Corvus corax among island habitats Canarian Archipelago. Ecoscience 6: 56–61.
Nogales M., Quilis V., Medina F.M., Mora J.L., Trigo L.S. (2002). Are predatory birds effective secondary seed dispersers? Biological Journal of the Linnean Society 75: 345–352.
Nogales M., Padilla D.P., Nieves C., Illera J.C., Traveset A. (2007). Secondary seed dispersal systems, frugivorous lizards and predatory birds in insular volcanic badlands. Journal of Ecology 95(6): 1394–1403. https://doi.org/10.1111/j.1365-2745.2007.01305.x.
Nogales M., Heleno R., Traveset A., Vargas P. (2012). Evidence for overlooked mechanisms of long-distance seed dispersal to and between oceanic islands. New Phytologist 194(2): 313–317. https://doi.org/10.1111/j.1469-8137.2011.04051.x.
Nybom H. (2004). Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13(5): 1143–1155. https://doi.org/10.1111/j.1365-294X.2004.02141.x.
Otto R., Krüsl B.O., Delgado J.D., Fernández-Palacios J.M., García Del Rey E., Arévalo J.R. (2010). Regeneration niche of the Canarian juniper: the role of adults, shrubs and environmental conditions. Annals of Forest Science 67(7): 709. https://doi.org/10.1051/forest/2010034.
Otto R., Barone R., Delgado J.D., Arévalo J.R., Garzón-Machado V., Cabrera-Rodríguez F., Fernández-Palacios J.M. (2012). Diversity and distribution of the last remnants of endemic juniper woodlands on Tenerife, Canary Islands. Biodiversity and Conservation 21(7): 1811–1824. https://doi.org/10.1007/s10531-012-0278-2.
Padilla D.P., González-Castro A., Nogales M. (2012). Significance and extent of secondary seed dispersal by predatory birds on oceanic islands: the case of the Canary archipelago. Journal of Ecology 100(2): 416–427. https://doi.org/10.1111/j.1365-2745.2011.01924.x.
Peakall R., Smouse P.E. (2006). Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6(1): 288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x.
Pritchard J.K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
Pritchard J.K., Wen W. (2004). Documentation for STRUCTURE software: version 2. University of Chicago, Chicago.
Quézel P., Pesson P. (1980). Biogeographie et ecologie des coniferes sur le pourtour mediterraneen. [Biogeography and ecology on the Mediterranean periphery]. In: Pesson P. (ed.). Actualites d’ecologie forestiere. [News on forest ecology]. Gauthier-Villars, Paris. p. 205–255.
Rivas-Martínez S., Wildpret M., del Arco M., Rodríguez O., Pérez-de-Paz P.L., García-Gallo A., Acebes J.R., Díaz-González T.E., Fernández-González F. (1993). Las comunidades vegetales de la Isla de Tenerife (Islas Canarias). [Plant communities from Tenerife (Canary Islands)]. Itinera Geobotanica 7: 169–374.
Rosenberg N.A. (2004). DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes 4(1): 137–138. https://doi.org/10.1046/j.1471-8286.2003.00566.x.
Rumeu B., Caujapé-Castells J., Blanco-Pastor J.L., Jaén-Molina R., Nogales M., Elias R.B., Vargas P. (2011). The colonization history of Juniperus brevifolia (Cupressaceae) in the Azores Islands. PLoS ONE 6(11): e27697. https://doi.org/10.1371/journal.pone.0027697.
Rumeu B., Vargas P., Jaén-Molina R., Nogales M., Caujapé-Castells J. (2014). Phylogeography and genetic structure of the threatened Canarian Juniperus cedrus (Cupressaceae). Botanical Journal of the Linnean Society 175(3): 376–394. https://doi.org/10.1111/boj.12172.
Sosa P.A., González- González E.A., González-Pérez M.A., Pérez-de-Paz P.L. (2013). Contrasting patterns of genetic differentiation in Macaronesian lineages of Ilex (Aquifoliaceae). Botanical Journal of the Linnean Society 173(2): 258–268. https://doi.org/10.1111/boj.12067.
Stuessy T.F., Jakubowsky G., Salguero-Gómez R., Pfosser M., Schluter P.M., Fer T., Sun B.-Y., Kato H. (2006). Anagenetic evolution in island plants. Journal of Biogeography 33(7): 1259–1265. https://doi.org/10.1111/j.1365-2699.2006.01504.x.
Stuessy T.F., Takayama K., López-Sepúlveda P., Crawford D.J. (2014). Interpretation of patterns of genetic variation in endemic plant species of oceanic islands. Botanical Journal of the Linnean Society 174(3): 276–288. https://doi.org/10.1111/boj.12088.
Su Y., Ting W., Deng F. (2010). Contrasting genetic variation and differentiation on Hainan Island and the Chinese mainland populations of Dacrycarpus imbricatus (Podocarpaceae). Biochemical Systematics and Ecology 38(4): 576–584. https://doi.org/10.1016/j.bse.2010.07.003.
Vargas P. (2007). Are Macaronesian Island refugia of relict plant lineages?: a molecular survey. In: Weiss S., Ferrand N. (eds.). Phylogeography of Southern European Refugia. Springer, Dortrecht, The Netherlands. p. 297–314. https://doi.org/10.1007/1-4020-4904-8_11.
Vargas P., Nogales M., Jaramillo P., Olesen J.M., Traveset A., Heleno R. (2014). Plant colonization across the Galápagos Islands: success of the sea dispersal síndrome. Botanical Journal of the Linnean Society 174(3): 349–358. https://doi.org/10.1111/boj.12142.
Vekemans X. (2002). AFLP-SURV version 1.0. Distributed by the author. Laboratoire de Génétique et Ecologie Végétale, Université Libre de Bruxelles, Bruxelles, Belgium. 15 p.
Vos P., Hogers R., Bleeker M., Reijans M., van de Lee T., Hornes M., Frijters A., Pot J., Peleman J., Kuiper M., Zabeau M. (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research 23(21): 4407–4414. https://doi.org/10.1093/nar/23.21.4407.
Whittaker R.J., Fernández-Palacios J.M. (2007). Island biogeography: ecology, evolution, and conservation. 2nd edn. Oxford University Press, Oxford.
Whittaker R.J., Triantis K.A., Ladle R.J. (2008). A general dynamic theory of oceanic island biogeography. Journal of Biogeography 35(6): 977–994. https://doi.org/10.1111/j.1365-2699.2008.01892.x.
Total of 73 references.

