Pollination dynamics variation in a Douglas-fir seed orchard as revealed by microsatellite analysis
Korecký J., El-Kassaby Y. A. (2016). Pollination dynamics variation in a Douglas-fir seed orchard as revealed by microsatellite analysis. Silva Fennica vol. 50 no. 4 article id 1682. https://doi.org/10.14214/sf.1682
Highlights
- Important characteristics such as parental reproductive success, pollen contamination, and selfing rate in the second generation Douglas-fir seed orchard have been estimated
- Since this research is a part of a multi-year study, outputs were compared to those from two other years
- Results are in line and differences in pollination dynamics across years are attributable to the various crop management practices.
Abstract
As part of a multi-year monitoring study of pollination dynamics in a second generation Douglas-fir (Pseudotsuga menziesii (Mirb.) Franco) seed orchard, we estimated with the aid of eight microsatellite markers three important reproductive biology characteristics affecting the genetic worth and diversity of seed crops; namely parental reproductive success, pollen contamination, and selfing rate. The obtained results were compared to those from two previous years to gauge appropriate seed crop management practices and ultimately allow approximate generalization of seed crop genetic quality. We determined that 80% of parental gametes were produced by 52% of the parents, 13% of paternal gametes resulted from pollen contamination (i.e., gene flow), and 12% of the seed were the product of selfing. The obtained results were in line with those observed for 2005 and 2009 where 80% of gametes being produced by 37–48% of the parents, 10–18% pollen contamination, and 15–17% selfing rate. The observed reproductive biology parameters differences are attributable to the various crop management practices implemented (i.e., bloom delay and supplemental-mass-pollination) across years and calls for justification due to the observed minimal differences on seed crops genetic quality.
Keywords
selfing;
Pseudotsuga menziesii (Mirb.) Franco;
genetic quality;
pedigree reconstruction;
gametic contribution;
pollen contamination
-
Korecký,
Department of Genetics and Physiology of Forest Trees, Faculty of Forestry and Wood Sciences, Czech University of Life Sciences Prague, Praha 6, 165 21, Czech Republic
 http://orcid.org/0000-0001-7859-1750
E-mail
korecky@fld.czu.cz
http://orcid.org/0000-0001-7859-1750
E-mail
korecky@fld.czu.cz

-
El-Kassaby,
Department of Forest and Conservation Sciences, Faculty of Forestry, University of British Columbia, Vancouver, BC, V6T 1Z4, Canada
 http://orcid.org/0000-0002-4887-8977
E-mail
y.el-kassaby@ubc.ca
http://orcid.org/0000-0002-4887-8977
E-mail
y.el-kassaby@ubc.ca
Received 4 August 2016 Accepted 9 September 2016 Published 15 September 2016
Views 105934
Available at https://doi.org/10.14214/sf.1682 | Download PDF

1 Introduction
Seed orchards link tree breeding and silviculture activities and are essential component of the forest tree improvement delivery system where gains from breeding are captured in their seed crops and delivered as genetically improved seedlings for reforestation activities (El-Kassaby 1995). Globally, seed orchards represent the main source of reforestation reproductive material (Funda and El-Kassaby 2012). Theoretically, seed orchard crops are expected to reflect their parental populations’ allelic frequencies, so gains attained through breeding, testing, and selection are reflected in their seedling crops. To reach this goal, seed orchards must function as a closed perfect panmictic populations (Eriksson et al. 1973). Practically, this goal is hardly achieved as a result of reproductive phenology asynchrony (El-Kassaby et al. 1984) and fertility variation (El-Kassaby et al. 1989) among the orchards’ parental population as well as extraneous pollen contamination from outside sources (i.e., gene flow) (Lai et al. 2010). In spite of these biological violations, seed orchards remained robust and successfully captured and delivered substantial genetic gains to reforestation programs (Funda and El-Kassaby 2012). To enhance the genetic quality of seed orchards’ crops, two management practices techniques; namely, supplemental mass pollination (SMP) (Silen and Keane 1969) and bloom delay (Wakeley et al. 1966) were commonly implemented in the Pacific Northwest. Supplemental mass pollination (SMP) involves the broadcast application of viable pollen to receptive and non-isolated female strobili in order to increase the seed set and the incorporation of desirable paternal gametic representation in seed crops (El-Kassaby and Davidson 1990; El-Kassaby et al. 1990). Bloom delay involves the application of an overhead fine water mist during late winter and early spring to slow the heat sum accumulation in order to postpone reproductive phenology process inside the seed orchard and thereby place its population out of synchrony with the ambient pollen and hence reducing pollen contamination (El-Kassaby and Ritland 1986a,b).
At their establishment, most first generation seed orchards’ designs judicially aimed at attaining equal parental representation (i.e., parents (clones) are represented by the same number of individuals (ramets) – see Faulkner 1975, for review). Initially, equal parental representation is thought to yield the same gametic contribution of the seed orchard parents, thus reflecting their allelic frequencies and therefore fulfilling fundamental quantitative genetics assumption of traits’ transmission over generations (El-Kassaby and Sziklai 1981). It should be stated that orchards’ equal parental representation assumes reproductive output equality among all parents (male and female gametes), a highly unattainable attribute due to its extreme variability (El-Kassaby et al. 1989; Moriguchi et al. 2004). After the completion of every breeding cycle, the genetic worth (breeding value) of the entire breeding population is estimated and subsequently a logical shift from parental equal representation to linear deployment was advocated by Lindgren and his co-workers (Lindgren and Matheson 1986; Bondesson and Lindgren 1993; Lindgren et al. 2009). The linear deployment concept proposes the use of different parental representation that is proportionate to their respective breeding values (i.e., high breeding value parents are represented with more individuals than their lower breeding value counterparts). Notably, the linear deployment concept also assumes that the gametic contribution is representative of the orchard’s parental linear representation. Finally, irrespective of which breeding strategy is implemented or seed orchard design is used, the main goal of the tree improvement endeavour is to reach an ideal balance between genetic gain and genetic diversity (Chaisurisri and El-Kassaby 1994; El-Kassaby and Ritland 1996).
The utility of genetic markers in studying seed orchard populations made it possible to account for parental actual gametic contribution to the seed crop and estimate levels of pollen contamination. When the actual gametic contribution is known, then seed crops’ genetic worth (represented by average breeding values weighted by parental gametic contribution) and genetic diversity (represented by effective population size) are precisely estimated (Stoehr et al. 1998; Slavov et al. 2005; El-Kassaby et al. 2010). Gene flow within seed orchard populations is affected by three main factors; namely, 1) females’ phenological asynchrony and males’ fertility variation, 2) level of gene flow from external pollen sources, and 3) level of self-fertilization (Burczyk et al. 2002). Variation in parental gametic contributions as well as gene flow produce unpredictable estimates of genetic gain and diversity. Thus, knowledge of both parental (trees in seed orchard) and offspring (seeds) gametic composition would enable adjustments of actual gain and diversity estimates as they often deviate from theoretical expectations. It is also essential to emphasize that the seed orchard parental population – seed crop analysis are year-specific and do not enable precise long-term predictions due to the commonly observed male and female reproductive success variation (El-Kassaby and Barclay 1992; Lai et al. 2010). Precise estimates of temporal variation of parental gametic contribution, gene flow, and selfing derived from genetic markers allow generalizations and provide support to practical on-ground survey-type assessment methods (Stoehr et al. 2004; Funda et al. 2011). Funda et al. (2014) presented an augment supporting the use of bulk seed samples coupled with the implementation of full pedigree reconstruction in determining the parental gametic profile of seed crops. Their hypotheses that random seed samples provide better representation of the parental (paternal and paternal) population and its pollination dynamics as opposed to the family-array methods that is spatially restricted to the seed-donors locations (Funda et al. 2014; El-Kassaby et al. 2015).
As a part of multi-year study aimed at investigating the pollination dynamics in a Douglas-fir (Pseudotsuga menziesii (Mirb.) Franco) seed orchard, this study was conducted to provide the information needed for 2007 seed-crop year. We used microsatellite markers analysis of a random bulk seed sample and implemented full-pedigree reconstruction to enable estimating parental reproductive success, selfing rate, and gene flow from non-orchard sources. The derived estimates are compared to those produced for 2005 (Lai et al. 2010) and 2009 (Kess and El-Kassaby 2015) seed crops with the goal of providing more fundamental insight into pollination dynamics processes including their annual variability and therefore develop informed recommendations for effective seed orchards’ crop management.
2 Materials and methods
2.1 Study site
A second-generation clonal Douglas-fir seed orchard located on southern Vancouver Island, British Columbia, Canada (48°35´N, 123°24´W, 50 m a.s.l.) provided the seed for this study. The seed orchard was established in 1990 using the permutated neighborhood design (Bell and Fletcher 1978) to minimize inbreeding. The parental population of the studied seed crop (2007) consisted of 57 parents and seed crop management included supplemental mass pollination (SMP) application with 21 within-orchard and 2 external paternal pollen-donors. SMP success rate was determined exclusively using the non-orchard pollen donors as within-orchard donors contributed to the orchard’s ambient pollination. According to the pedigree records of British Columbia Ministry of Forests’ (Michael Stoehr, personal communication in 2015) the seed orchard harbored some level of co-ancestry with two parents as members of a single full-sib family and four cases of parent-offspring.
2.2 DNA extraction, PCR amplification and genotyping
DNA from the parental and offspring populations was extracted from young vegetative buds and a random sample of bulk seed (2007 crop), respectively. A total of 402 germinated seeds were dissected into diploid (2n) embryo and their corresponding maternal haploid (1n) megagametophyte using the CTAB extraction protocol of Doyle (1990). The weight of these seeds before germination was 4.55 grams in total, the weight of the entire seed crop of the seed orchard was estimated to be 19.3 kg. All PCR reactions contained 25 ng of template DNA, 0.25 mM of each dNTP, 1x Amplitaq Gold PCR buffer (Thermo Fisher Scientific, Waltham, MA) and 1 mM MgCl2 and 0.25 µl forward and 0.25 µl reverse primer in final volume of 10 µl.
A battery of eight Douglas-fir microsatellite primers (PmOSU_2G12, PmOSU_3B9, PmOSU_3F1, PmOSU_4G2, PmOSU_3D5, PmOSU_2D4, PmOSU_3G9 and PmOSU_2C2) developed by Slavov et al. (2004) was used for DNA fingerprinting. PCR reaction conditions, amplified in a GeneAmp 9700 thermal cycler and an Eppendorf Mastercycler gradient thermal cycler, followed those reported by Slavov et al. (2004) and DNA amplicons were separated on polyacrylamide gels using LiCor 4200 automated sequencer (LiCor Inc., Lincoln, NE) and genotyped by SAGATM software (LiCor Inc., Lincoln, NE) followed by final manual adjustment.
2.3 Data analysis and pedigree reconstruction
The maternal parent was determined by comparison of multi-locus megagametophyte genotype with genotype of all potential maternal trees using a program written in Microsoft VisualBasic® (Lai et al. 2010). Mismatches between maternal parent and assigned offspring at a particular locus (most likely arising from null alleles) were manually corrected. The genetic parameters were calculated using GenAlEx 6.5 (Peakall and Smouse 2012) and paternity analysis was done using the CERVUS program (Kalinowski et al. 2007) with 95% assignment probability and allowance for 0.01 error rate, self-fertilization, and mutation. To secure that the same input parameters were used across seed crops for different years, the proportion of genotyped paternal population (Nbmp) was set to 0.95 (Lai et al. 2010; Kess and El-Kassaby 2015). Simulation using the eight markers indicated that under these parameters 0.99 of the 10 000 simulated offspring could be assigned to parents. Genetic diversity represented by effective population size (Ne) was estimated using the concept of molecular co-ancestry (Nomura 2008) which leads to the precise estimation of the current or so called short-term Ne (Wang 2005) implemented in the software NeEstimator v2 (Do et al. 2014). Estimates of correlation between reproductive energy (male and female cones) and reproductive success (realized gametic contribution based on genotyping results) were generated using software Statistica 12® and Microsoft Excel. Maternal reproductive energy has been estimated as proportion of collected cones for each clone to total cones volume and for paternal reproductive energy in similar way via pollen count. In general, orchard‘s male gamete contribution is determined using the number of male-cone (pollen bud) clusters for each ramet and assigning a crop intensity rating for this tree (i.e., 100% sampling). The categorical ratings are very heavy, heavy, medium, light, and very light which subsequently are converted into a quantitative estimate of male reproductive output Female gamete contributions for each clone are estimated using either the seed-cone volume or the weight of cones produced.
3 Results
The combined non-exclusion probability of the one candidate parent given the genotype of a known parent for all loci is 1.01 × 10–5 indicating high robustness of the selected loci for parentage assignment. Mean polymorphic information content (PIC: Botstein et al. 1980) across the eight studied loci was 0.857 ± 0.019 (SE) (Table 1). A seemingly high estimate of null alleles was presumed 0.232 ± 0.035 (Table 1). Nevertheless more important than the estimator itself, which does not actually estimate the level of null alleles rather than indicate the deviation between expected and observed heterozygosities under H-W expectations, is the null alleles occurrence across all loci which is further discussed below. Generally, the number of alleles per locus varied between the orchard’s parental and offspring populations (17.6 vs. 22.1) indicating effective gametic intrusion (gene flow/contamination) from the non-orchard sources (Table 1). One exception case where the number of alleles per locus was higher in parents compared to offspring was observed at the Pm_OSU3D5 locus, indicating that the sampled seed did not capture the parental population’s allelic profile (two missing alleles) (Table 1). A slight and not significant decrease in the observed heterozygosity was observed between the parents and their offspring (0.562 vs. 0.556) (Table 1). The effective population size estimate based on molecular co-ancestry method (proportion of parents involved in the production of the seed crop) was 40.2, a relatively high estimate considering a census number of 55 parents.
| Table 1. Genetic diversity of seed orchard parents and offspring. | ||||||||
| Marker | Parents | Offspring | ||||||
| Locus | PIC | F(null) | A | Ho | He | A | Ho | He |
| 2G12 | 0.855 | 0.154 | 15.00 | 0.438 | 0.858 | 19.00 | 0.665 | 0.867 |
| 3B9 | 0.860 | 0.165 | 19.00 | 0.636 | 0.900 | 27.00 | 0.624 | 0.863 |
| 3F1 | 0.912 | 0.225 | 21.00 | 0.630 | 0.928 | 25.00 | 0.574 | 0.917 |
| 4G2 | 0.855 | 0.096 | 10.00 | 0.776 | 0.863 | 14.00 | 0.706 | 0.870 |
| 3D5 | 0.862 | 0.288 | 18.00 | 0.419 | 0.887 | 16.00 | 0.500 | 0.873 |
| 2D4 | 0.937 | 0.180 | 26.00 | 0.510 | 0.953 | 30.00 | 0.681 | 0.938 |
| 3G9 | 0.840 | 0.419 | 17.00 | 0.436 | 0.861 | 26.00 | 0.342 | 0.851 |
| 2C2 | 0.738 | 0.326 | 15.00 | 0.647 | 0.877 | 20.00 | 0.358 | 0.745 |
| Mean | 0.857 | 0.232 | 17.63 | 0.562 | 0.891 | 22.13 | 0.556 | 0.866 |
| SE | 0.019 | 0.035 | 1.56 | 0.043 | 0.011 | 1.88 | 0.047 | 0.019 |
| PIC – polymorphic information content F(null) – null allele frequency estimator A – allelic richness Ho – observed heterozygosity He – expected heterozygosity | ||||||||
Allelic concordance between alleged maternal parents and their putative progeny (N = 402) were compared. We found low proportion of genotypic mismatches (0.0064 across all loci) which indicate no maternal contaminants (i.e., seed mixing from other sources) and low occurrence of null alleles. If partial mother-offspring mismatches at one or two loci could be explained by non-amplifying allele (apparent incorrect homozygous maternal or offspring genotype), genotypes were then corrected to include this missing allele. After this adjustment low but still remaining proportion of allelic mismatches might be explained by mutation process between generations (Jones and Ardren 2003).
A 13.18% (53 individuals) gametic contribution of non-orchard pollen sources (i.e., gene flow/pollen contamination) was estimated. Of the 349 individual seed pollinated by within orchard pollen were assigned to both parents, of which 42 resulted from selfing, producing a selfing rate of 12.04%. Based on the two non-orchard pollen donors, the SMP success rate (i.e., gametic contribution from outside pollen donors) was only 0.57% (2 seeds from the subset of 349 individuals with the assigned fathers).
Maternal, paternal, and parental reproductive success varied among the orchard’s parents and all were lower than the ideal scenario of equal contribution and indicating the presence of greater variation in female reproductive success compared to their male counterparts (Fig. 1). Considering gametic contribution of 0.8 as a benchmark, maternal, paternal, and parental contributed 0.393 (22 individuals), 0.554 (31), and 0.518 (29), respectively (Fig. 1). Male gametic contribution slightly mirrored that of the female with a significant (p < 0.01) Pearson product-moment correlation coefficient of r = 0.519. However, while correlated, female and male reproductive success followed different trajectories as indicated by the coefficient of determination (R2 = 0.269) indicating the presence of different male and female reproductive successes. It is important to point out that the use of randomly drawn bulk sample of seed as a representative of the entire orchard’s seed crop enabled the unbiased estimation of female reproductive success as opposed to the commonly practiced sampling of equal number of seed from a subset of the orchard’s parents which is only capable of estimating male rather than female reproductive success.
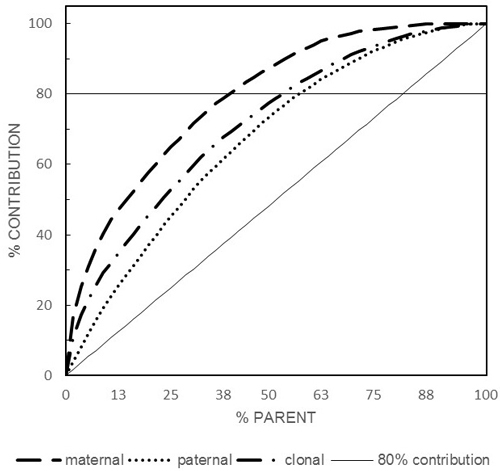
Fig. 1. Paternal, maternal and clonal cumulative reproductive success (80% contribution is represented by the horizontal line).
Pearson product moment correlation between female reproductive investment represented by the volume of harvested cones and female reproductive success determined from the gametic contribution was relatively high correlation (r = 0.868; p < 0.01). Similarly, the correlation between male reproductive investment assessed by pollen count and reproductive success determined from gametic contribution significant (r = 0.482; p < 0.01)), yet lower that observed for female. These results confirmed earlier observations on the approximate relationship between on-ground reproductive investment surveys and DNA-based estimates and highlighting the strength of this relationship for female rather than male reproductive success.
The assigned paternal and maternal gametic contributions were used to construct a three dimensional graph depicting the mating pattern across the seed orchard’s parents (Fig. 2). Each column represents a full-sib family (i.e., a cross between particular maternal and paternal parents) with the largest full-sib of sample size of 6 progenies (Fig. 2).
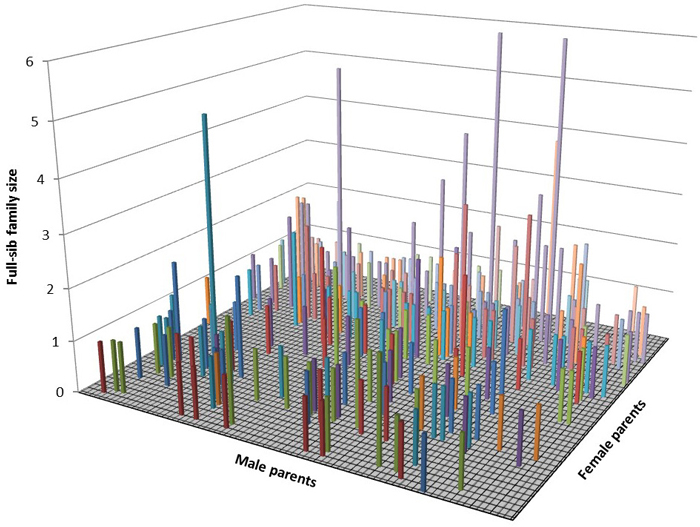
Fig. 2. Three dimensional representation of the mating dynamics in the seed orchard.
4 Discussion
The observed results from the present seed crop (2007) were in line with that obtained from 2005 (Lai et al. 2010) and 2009 (Kess and El-Kassaby 2015) seed crops, indicating somewhat consistent outcome over time. Although this study was conducted as a part of a multi-year assessment of a single seed orchard it should be noted that the seed orchard population is in the dynamic state across studied years due to the removal and inclusion of the reproduction participating parents. Even this minor variation in contributing parents among years is expected to have an influence on parental gametic composition. Therefore, comparing the dynamics and changes of reproductive biology parameters across the three studied years’ (2005, 2007 and 2009) seed crops would be useful and informative.
Evaluating the genetic diversity of the parental and offspring populations considering the 4 shared loci among the studied years (i.e., 2G12, 3F1, 2D4 and 3G9), we detected different observed heterozygosity (Ho) values. Parental population’s heterozygosity estimates values of 0.684, 0.504, and 0.727 where observed for 2005 (Lai et al. 2010), 2007 (present study), and 2009 (Kess and El-Kassaby 2015), respectively. We attribute the cause of the observed difference is mainly as a result of the dynamic reproductive state of the seed orchard’s parental population. For example, the number of parents contributed to the seed crop were 49, 57, and 66 for 2005, 2007, and 2009 seed crop, respectively. Additionally, in 2009 the parental population increased by 13.6% as the result of incorporating new parents. New clones are entered the seed orchard in any year as ramets growing in large pots, thus they are of reproductive maturity. It is also interesting to note that the observed heterozygosity of the offspring populations were balanced values among years with 0.721, 0.566, and 0.665 for 2005, 2007, 2009, respectively.
The allele frequency analysis processed by Cervus showed the potential presence of null alleles in some parents for a set of utilized loci. Nevertheless it is important to point out, that it provided information on the heterozygote deficit relative to Hardy-Weinberg equilibrium, that might be caused not only by the occurrence of null alleles (e.g., Jarne and Lagoda 1996) but also by other biological factors such as relatedness or non-random mating among the individuals (Dakin and Avise 2004). We believe that the indicator of potential presence of null alleles could be explained by the biological factors mentioned above, as we can formulate certain assumptions about the kinship among the parental generation (see Material and Methods section) and we found the occurrence of potential null alleles more or less concordant across all analyzed loci (SE 0.035), whereas the effects of null alleles are locus-specific. Indeed Slavov et al. (2004), who developed these markers, evaluated them as having low occurrence of null alleles (0.062 in their dataset). They also confirmed their Mendelian inheritance and even their dispersion throughout the genome. We used the approach proposed by Slavov et al. (2005), namely to compare genotype of maternally contributed haplotype and the diploid genotype of the embryo for the obvious mismatches. Although it is not possible to reveal all sources of mistyping (Ewen et al. 2000), this approach indicates the proportion of null alleles more precisely than that based only on the deviation from H-W equilibrium, without assumptions about the biological nature of the data.
The observed frequency of obvious mismatches in maternal parent ranged from 0.0029 (1 maternal – embryogenic mismatch) to 0.0117 (inconsistency of 5 maternal – embryogenic genotypes) among loci with mean of 0.0064. The high discrepancy between the estimator of potential null alleles occurrence obtained by Cervus (0.232 ± 0.035) and frequencies of mismatches (i.e., the explanatory proportion of null alleles) are also in concordance with our hypothesis about the biological composition (i.e., relatedness among seed orchard parents and thus the deviation from H-W expectations of homozygotes/heterozygotes distribution).
The sample set of seed was collected randomly from the entire seed crop after proper mixing. It has been reported that a sample size of 30 individuals/seed is adequate to capture alleles frequency of < 0.05 in the population (El-Kassaby 1991). Thus the randomly drawn bulk sample of 402 seed is assumed to provide a reasonable representation of both maternal and paternal reproductive output and thus enable the elucidation of unbiased inferences on both female and male reproductive biology attributes such as parental gametic contribution (reproductive success) and contamination and selfing rates (Lai et al. 2010).
We feel that the results from these three years were somewhat consistent as 80% of the gametic contribution was attributable to 37–52% of the parental population, with pollen contamination rate ranging between 10 and 18%, and selfing rate ranging between 12 and 17% (Lai et al. 2010; Kess and El-Kassaby 2015; the present study). We would like to state that our contamination estimate represents the so called apparent pollen flow (Devlin and Ellstrand 1990) and does not reflect the portion of external pollen flow known as “cryptic gene flow” which is often missed due to similarity between the genotype of the contaminating pollen and the inside the orchard maternal parents. Our approach is similar to that implemented by Slavov et al. (2005), Adams et al. (1997), and Burczyk and Prat (1997) in other Douglas-fir pollen contamination studies in which cryptic gene flow was also undetermined.
As indicated in the M&M section, for the sake of consistency across the three studied crops (years) we set the assumed proportion of genotyped males (Nbmp) at 0.95. However, Oddou-Muratorio et al. (2003) suggested that the software PATRI (Nielsen et al. 2001) is better suited to obtaining more accurate estimation of Nbmp as an input parameter for analysis in Cervus. We did not implement this approach as PATRI is not effective with missing data as in our case where missing data were observed across the studied loci and individuals.
Among the three studies there are greater differences between maternal reproductive success compared to their paternal counterpart considering the 80% threshold (2005: 0.230 vs. 0.310, 2007: 0.393 vs. 0.554, and 2009: 0.394 vs. 0.454 for paternal vs. maternal, respectively), indicating that individuals’ reproductive success is a seed crop-specific and generalization is better done on total parental reproductive success rather than either paternal or maternal. Additionally, the seed crop management practices implemented as well as the orchard’s parental status (i.e., number of parents/clones) could have influenced the observed reproductive biology parameters; however, the observed differences are not that drastic to warrant a specific consideration. For example: 1) the 2005 seed crop was managed under bloom delay treatment while the other two years were under ambient condition, 2) the orchard’s population was in a constant change as the number of parents kept increasing overtime from 49 to 57 to 66 for 2005, 2007, and 2009, respectively, 3) each seed-crop year is characterized by its own reproductive phenology chorography, thus influencing the mating pattern among the orchard’s parents as well as creating different opportunities for gene flow (Fashler and El-Kassaby 1987), and 4) varying seed-cone crop size among the studied year (2005: 0.151 kg.hL–1 (small), 2007: 843 kg.hL–1 (medium), 2009: 2599 kg.hL–1 (large) crops). Generally, while the reproductive biology parameters slightly varied among the studied years, mainly due to the factors mentioned above, our results are in agreement of those reported by Slavov et al. (2005) for another Douglas-fir seed orchard studied over a similar three-year period with only a notable exception that is related to the level of gene flow which was 0.353 compared to our mean of 0.140. The high rate of pollen contamination reported for Slavov et al. (2005) seed orchard also affected the apparent selfing rate as it is expected that higher pollen contamination often lead to lower selfing as each pollen contamination event is an outcrossing event (El-Kassaby and Ritland 1986a,b).
5 Conclusion
Highly polymorphic microsatellite markers allowed analysis of reproductive biology dynamics within the seed orchard. Gametic contributions, as expected, deviated from the theoretical expectations of panmixia. Pedigree reconstruction allowed estimation of important reproductive biology parameters of biological and operational importance such as parental gametic contribution and gene flow and selfing rates. The analysis of pollination dynamics in the studied orchard produced somewhat comparable results, encouraging outcome as the orchard’s population itself was in a dynamic state caused by the removal and introduction of new parents over time as well as the application of different seed crop management practices such as bloom delay (2005) and SMP over the studied years.
Acknowledgements
Funds from the Johnson’s Family Forest Biotechnology Endowment, the Natural Sciences and Engineering Research Council of Canada – Discovery grant to YAK and the University-wide Internal Grant Agency of the Czech University of Life Sciences (CIGA; no. 20144301) to JK are greatly appreciated.
References
Adams W.T., Hipkins V.D., Burczyk J, Randall W.K. (1997). Pollen contamination trends in a maturing Douglas-fir seed orchard. Canadian Journal of Forest Research 27(1): 131–134. http://dx.doi.org/10.1139/x96-129.
Bell G.D., Fletcher A.M. (1978). Computer organised orchard layouts (COOL) based on the permutated neighbourhood design concept. Silvae Genetica 27: 223–225.
Bondesson L., Lindgren D. (1993). Optimal utilization of clones and genetic thinning of ’seed orchards. Silvae Genetica 42(4–5): 157–163.
Botstein D., White R.L., Skolnick M., Davis R.W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics 32: 314–31.
Burczyk J., Prat D. (1997). Male reproductive success in Pseudotsuga menziesii (Mirb.) Franco: the effects of spatial structure and flowering characteristics. Heredity 79: 638–647. http://dx.doi.org/10.1038/hdy.1997.210.
Burczyk J., Adams W.T., Moran G.F., Griffin A.R. (2002). Complex patterns of mating revealed in a Eucalyptus regnans seed orchard using allozyme markers and the neighbourhood model. Molecular Ecology 11(11): 2379–2391. http://dx.doi.org/10.1046/j.1365-294X.2002.01603.x.
Chaisurisri K., El-Kassaby Y.A. (1994). Genetic diversity in a seed production population vs. natural populations of Sitka Spruce. Biodiversity and Conservation 3(6): 512–523. http://dx.doi.org/10.1007/BF00115157.
Dakin E.E., Avise J.C. (2004). Microsatellite null alleles in parentage analysis. Heredity 93: 504–509. http://dx.doi.org/10.1038/sj.hdy.6800545.
Devlin B., Ellstrand N.C. (1990). The development and application of a refined method for estimating gene flow from angiosperm paternity analysis. Evolution 44: 248–259. http://dx.doi.org/10.2307/2409404.
Do C., Waples R.S., Peel D., Macbeth G.M., Tillett B.J., Ovenden J.R. (2014). NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Molecular Ecology Resources 14(1): 209–214. http://dx.doi.org/10.1111/1755-0998.12157.
Doyle J., Doyle J. (1990). Isolation of plant DNA from fresh tissue. Focus 12: 13–15.
El-Kassaby Y.A. (1991). Genetic variation within and among conifer populations: review and evaluation of methods. In: Fineschi S, Malvolti M.E., Cannata F., Hattermer H.H. (eds.). Biochemical markers in the population genetics of forest trees. SPB Academic Publishing, The Hague, The Netherlands. p. 61–76.
El-Kassaby Y.A. (1995). Evaluation of the tree-improvement delivery system: factors affecting genetic potential. Tree Physiology 15: 545–550. http://dx.doi.org/10.1093/treephys/15.7-8.545.
El-Kassaby Y.A., Barclay H.J. (1992). Cost of reproduction in Douglas fir. Canadian Journal of Botany 70: 1429–1432. http://dx.doi.org/10.1139/b92-179.
El-Kassaby Y.A., Davidson R. (1990). Impact of crop management practices on the seed crop genetic quality in a Douglas-fir seed orchard. Silvae Genetica 39: 230–237.
El-Kassaby Y.A., Ritland K. (1986a). The relation of outcrossing and contamination to reproductive phenology and supplemental mass pollination in a Douglas-fir orchard. Silvae Genetica 35: 240–244.
El-Kassaby Y.A., Ritland K. (1986b). Low levels of pollen contamination in a Douglas-fir seed orchard as detected by allozyme markers. Silvae Genetica 35: 224–229.
El-Kassaby Y.A., Ritland K. (1996). Impact of selection and breeding on the genetic diversity in Douglas-fir. Biodiversity and Conservation 5: 795–813. http://dx.doi.org/10.1007/bf00051787.
El-Kassaby Y.A., Sziklai O. (1982). Genetic variation of allozyme and quantitative traits in a selected Douglas-fir [Pseudotsuga menziesii var. menziesii (Mirb.) Franco] population. Forest Ecology and Management 4(2): 115–126. http://dx.doi.org/10.1016/0378-1127(82)90009-3.
El-Kassaby Y.A., Fashler A.M.K., Sziklai O. (1984). Reproductive phenology and its impact on genetically improved seed production in a Douglas-fir seed orchard. Silvae Genetica 33: 120–125.
El-Kassaby Y.A., Fashler A.M.K., Crown M. (1989). Variation in fruitfulness in a Douglas-fir seed orchard and its effect on crop-management decisions. Silvae Genetica 38: 113–121.
El-Kassaby Y.A., Edwards D.G.W., Cook C. (1990). Impact of crop management practices on seed yield in a Douglas-fir seed orchard. Silvae Genetica 39: 226–230.
El-Kassaby Y.A., Funda T., Lai B.S.K. (2010). Female reproductive success variation in a pseudotsuga menziesii seed orchard as revealed by pedigree reconstruction from a bulk seed collection. Journal of Heredity 101: 164–168. http://dx.doi.org/10.1093/jhered/esp126.
El-Kassaby Y.A., Funda T., Liewlaksaneeyanawin C. (2015). Increasing Breeding without Breeding (BwB) efficiency: full-vs. partial-pedigree reconstruction in Lodgepole pine. SOJ Genetic Science 2: 1–6.
Eriksson G., Lindgren D., Jonsson A. (1973). Flowering in a clone trial of Picea abies Karst. Studia Forestalia Suecica 110: 3–45.
Ewen K.R., Bahlo M., Treloar S.A., Levinson D.F., Mowry B., Barlow J.W., Foote S.J. (2000). Identification and analysis of error types in high-throughput genotyping. American Journal of Human Genetics 67(3): 727–736. http://dx.doi.org/10.1086/303048.
Fashler A.M.K., El-Kassaby Y.A. (1987).The effect of water spray cooling treatment on reproductive phenology in a Douglas-fir seed orchard. Silvae Genetica 36: 245–249.
Faulkner R. (1975). Seed orchards. Forestry Commision Bulletin 54. 149 p.
Funda T., El-Kassaby Y.A. (2012). Seed orchard genetics. CAB reviews: perspectives in agriculture, veterinary science, nutrition and natural resources 7(13). 23 p.
Funda T., Liewlaksaneeyanawin C., Fundova I., Lai B.S.K., Walsh C., Van Niejenhuis A., Cook C., Graham H., Woods J., El-Kassabyb Y.A. (2011). Congruence between parental reproductive investment and success determined by DNA-based pedigree reconstruction in conifer seed orchards. Canadian Journal of Forest Research 41(2): 380–389. http://dx.doi.org/10.1139/x10-190.
Funda T., Liewlaksaneeyanawin C., El-Kassaby Y.A. (2014). Determination of paternal and maternal parentage in lodgepole pine seed: full versus partial pedigree reconstruction. Canadian Journal of Forest Research 44: 1122–1127. http://dx.doi.org/10.1139/cjfr-2014-0145.
Jarne P., Lagoda P.J.L. (1996). Microsatellites, from molecules to populations and back. Trends in ecology and evolution 11: 424–429. http://dx.doi.org/10.1016/0169-5347(96)10049-5.
Jones A.G., Ardren W.R. (2003). Methods of parentage analysis in natural populations. Molecular Ecology 12: 2511–2523. http://dx.doi.org/10.1046/j.1365-294X.2003.01928.x.
Kalinowski S.T., Taper M.L., Marshall T.C. (2007). Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Molecular Ecology 16: 1099–1106. http://dx.doi.org/10.1111/j.1365-294X.2007.03089.x.
Kess T., El-Kassaby Y.A. (2015). Estimates of pollen contamination and selfing in a coastal Douglas-fir seed orchard. Scandinavian Journal of Forest Research 30(4): 266–275. http://dx.doi.org/10.1080/02827581.2015.1012112.
Lai B.S., Funda T., Liewlaksaneeyanawin C., Klápště J., Van Niejenhuis A., Cook C., Stoehr M.U., Woods J., El-Kassaby Y.A. (2010). Pollination dynamics in a Douglas-fir seed orchard as revealed by pedigree reconstruction. Annals of Forest Science 67(8): 808–808. http://dx.doi.org/10.1051/forest/2010044.
Lindgren D., Danusevicius D., Rosvall O. (2009). Unequal deployment of clones to seed orchards by considering genetic gain, relatedness and gene diversity. Forestry 82: 17–28. http://dx.doi.org/10.1093/forestry/cpn033.
Lindgren D., Matheson A.C. (1986). An algorithm for increasing the genetic quality of seed from seed orchards by using the better clones in higher proportions. Silvae Genetica 35: 173–177.
Moriguchi Y., Taira H., Tani N., Tsumura Y. (2004). Variation of paternal contribution in a seed orchard of Cryptomeria japonica determined using microsatellite markers. Canadian Journal of Forest Research 34: 1683–1690. http://dx.doi.org/10.1139/X04-029.
Nielsen R., Mattila D.K., Clapham P.J., Palsboll P.J. (2001). Statistical approaches to paternity analysis in natural populations and applications to the North Atlantic humpback whale. Genetics 157: 1673–1682.
Nomura T. (2008). Estimation of effective number of breeders from molecular coancestry of single cohort sample. Evolutionary Applications 1(3): 462–474. http://dx.doi.org/10.1111/j.1752-4571.2008.00015.x.
Oddou-Muratorio S., Houot M.L., Demesure-Musch B., Austerlitz F. (2003). Pollen flow in the wildservice tree, Sorbus torminalis (L.) Crantz. I. Evaluating the paternity analysis procedure in continuous populations. Molecular Ecology 12: 3427–3439. http://dx.doi.org/10.1046/j.1365-294X.2003.01989.x.
Peakall R., Smouse P.E. (2012). GenALEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28: 2537–2539. http://dx.doi.org/10.1093/bioinformatics/bts460.
Silen R., Keane G. (1969). Cooling a Douglas-fir seed orchard to avoid pollen contamination. Note PNW-101, USDA Forestry Service.
Slavov G.T., Howe G.T., Adams W.T. (2005). Pollen contamination and mating patterns in a Douglas-fir seed orchard as measured by simple sequence repeat markers. Canadian Journal of Forest Research 35: 1592–1603. http://dx.doi.org/10.1139/x05-082.
Slavov G.T., Howe G.T., Yakovlev I., Edwards K.J., Krutovskii K.V., Tuskan G.A., Carlson J.E., Strauss S.H., Adams W.T. (2004). Highly variable SSR markers in Douglas-fir: Mendelian inheritance and map locations. Theoretical and Applied Genetics 108(5): 873–880. http://dx.doi.org/10.1007/s00122-003-1490-y.
Stoehr M., Webber J., Woods J. (2004). Protocol for rating seed orchard seedlots in British Columbia: quantifying genetic gain and diversity. Forestry 77(4): 297–303. http://dx.doi.org/10.1093/forestry/77.4.297.
Stoehr M.U., Vo T.M., Gawley J.R., Webber J.E., Newton C.H. (1998). Application of a chloroplast DNA marker in seed orchard management evaluations of Douglas-fir. Canadian Journal of Forest Research 28(2): 187–195. http://dx.doi.org/10.1139/cjfr-28-2-187.
Wakeley P.C., Wells O.O., Campbell T.E. (1966). Mass production of shortleaf x slash pine hybrids by pollinating unbagged female flowers. Joint proceedings of the second genetics workshop of the Society of American Foresters and the Seventh Lake States Forest Tree Improvement conference. USDA Forest Service, St. Paul, USA. p 78–79.
Wang J. (2005). Estimation of effective population sizes from data on genetic markers. Philosophical Transactions of the Royal Society B, Biological Sciences 360(1459): 1395–1409. http://dx.doi.org/10.1098/rstb.2005.1682.
Total of 50 references.

