Metagenomic approach of associated fungi with Megaplatypus mutatus (Coleoptera: Platypodinae)
Ceriani-Nakamurakare E., Ramos S., Robles C. A., Novas M. V., D´Jonsiles M. F., Gonzalez-Audino P., Carmarán C. (2018). Metagenomic approach of associated fungi with Megaplatypus mutatus (Coleoptera: Platypodinae). Silva Fennica vol. 52 no. 3 article id 9940. https://doi.org/10.14214/sf.9940
Highlights
- There were no significant effects of host plant and location on fungal richness
- Two fungal species, belonging to Fusarium and Candida genera, were present in all the studied associations
- Results suggest that host plant identity would not be crucial to determine the composition of fungal communities associated to Megaplatypus mutatus.
Abstract
Megaplatypus mutatus is a major forest pest in Argentina and an emerging pest in Europe. In this study the multitrophic interactions between M. mutatus and associated fungi were assessed with a metagenomics approach (454-pyrosequencing). A total of 270 collection points from insect galleries from three locations in Argentina were pooled for pyrosequencing analyses. Two hosts, Populus deltoides and Casuarina cunninghamiana, were independently evaluated to characterize the fungal communities associated to M. mutatus; compare the culture-independent approach with previous culturing studies, in terms of data recovery related to the fungal community composition, and test the specificity of the fungal communities amongst locations and hosts. A Generalized Linear Mixed Model was performed to compare the fungal richness in each dataset, which showed no significant differences between taxa richness amongst locations. Principal Coordinates Analyses showed a separation between fungal communities within the same host, suggesting that host identity would not be crucial to determine the specificity in fungal communities. Candida insectalens and one Fusarium species, present in all hosts and locations, achieved 37.6% of the total relative frequency per taxa. These results complement the data from culturing methods previously reported, thus improving the accuracy and understanding of the fungal assemblages associated to M. mutatus.
Keywords
ambrosia;
forest pest;
metagenomic;
fungi;
Platypodinae;
insect-fungus interactions
- Ceriani-Nakamurakare, Universidad de Buenos Aires. Facultad de Ciencias Exactas y Naturales. Depto. Biodiversidad y Biología Experimental. Buenos Aires, Argentina; CONICET- Universidad de Buenos Aires. Instituto de Micología y Botánica (INMIBO). Buenos Aires, (C1428EHA) Argentina E-mail cerianinaka@gmail.com
- Ramos, Instituto Nacional de Tecnología Agropecuaria. Estación Experimental Agropecuaria Concordia. Entre Ríos, (E3200) Argentina E-mail ramos.sergio@inta.gob.ar
- Robles, Universidad de Buenos Aires. Facultad de Ciencias Exactas y Naturales. Depto. Biodiversidad y Biología Experimental. Buenos Aires, Argentina; CONICET- Universidad de Buenos Aires. Instituto de Micología y Botánica (INMIBO). Buenos Aires, (C1428EHA) Argentina E-mail carorobles@bg.fcen.uba.ar
- Novas, Universidad de Buenos Aires. Facultad de Ciencias Exactas y Naturales. Depto. Biodiversidad y Biología Experimental. Buenos Aires, Argentina; CONICET- Universidad de Buenos Aires. Instituto de Micología y Botánica (INMIBO). Buenos Aires, (C1428EHA) Argentina E-mail vicnovas@bg.fcen.uba.ar
- D´Jonsiles, Universidad de Buenos Aires. Facultad de Ciencias Exactas y Naturales. Depto. Biodiversidad y Biología Experimental. Buenos Aires, Argentina; CONICET- Universidad de Buenos Aires. Instituto de Micología y Botánica (INMIBO). Buenos Aires, (C1428EHA) Argentina E-mail lalijonsi@gmail.com
- Gonzalez-Audino, Centro de Investigaciones de Plagas e Insecticidas (CITEFA-CONICET). Buenos Aires, (B1603ALO) Argentina E-mail pgonzalezaudino@citedef.gob.ar
-
Carmarán,
Centro de Investigaciones de Plagas e Insecticidas (CITEFA-CONICET). Buenos Aires, (B1603ALO) Argentina
E-mail
carmaran@bg.fcen.uba.ar

Received 28 December 2017 Accepted 9 May 2018 Published 15 May 2018
Views 121670
Available at https://doi.org/10.14214/sf.9940 | Download PDF

Supplementary Files
1 Introduction
Interactions established between insects and fungi have been more intensively studied in recent years, including those involving bark beetles (Hulcr and Stelinski 2016). Previous research has shown that the information provided could offer new tools for pest insect management (Castrillo et al. 2016). Megaplatypus mutatus (Chapuis) belongs to the Platypodinae subfamily, whose members along with Scolytinae, are known as ambrosia beetles (Coleoptera: Curculionidae). Unlike most bark and ambrosia beetles, M. mutatus attacks vigorous trees (Alfaro et al. 2007), where a single gallery built by only one couple of beetles weakens the trunk causing its breakage under the effect of strong wind (Supplementary file S1) (González-Audino et al. 2011; Bobrowsky 2013). Moreover, dark staining produced by the ambrosial mycelia growing on the gallery walls reduces the wood quality (Bascialli et al. 1996). Megaplatypus mutatus has a wide host range (Giménez and Etiennot 2007), it has been reported in several countries (Charles et al. 2014). A major forest pest in Argentina (Alfaro et al. 2007) has been considered in this study, achieving the status of an invasive species and an emerging non-native pest in Europe (Tremblay et al. 2000; Allegro and Della Beffa 2001; Funes et al. 2011). In Argentina, this pest has been found infesting several forest tree species. Investigations of two tree species with commercial importance are included in this study: Populus deltoides W. Bartram ex Marshall and Casuarina cunninghamiana Miq. (Giménez and Etiennot 2007).
The first studies about ambrosia interactions were based on the specificity between a beetle and a single dominant fungus (Francke-Grosmann 1956; Batra 1963). However, an increasing number of studies suggest that certain beetle species can have multiple fungal associates (Batra 1966; Gebhardt et al. 2004; Ceriani-Nakamurakare et al. 2016). Moreover, Kostovcik et al. (2015) suggested that the ambrosia interaction is ecologically dynamic and more species-rich than any other insect–fungus interaction. Ceriani-Nakamurakare et al. (2016) characterized the fungal community associated with M. mutatus and Populus deltoides based on culture dependent methods. Those results showed a high fungal diversity that included the ambrosia fungi, Raffaelea spp. Also other fungal species were frequently detected as having various ecological roles, members of the Fusarium solani and F. oxysporum species complexes, Graphium basitruncatrum, Coprinellus radians and members of Dipodascaceae family were most commonly found. These taxa represented 94.15% of the fungal species isolated from galleries of M. mutatus highlighting the presence of complex, multitrophic interactions (Ceriani-Nakamurakare et al. 2016).
High-throughput sequencing technologies have improved the understanding of true microbial diversity, as large amounts of genetic information can be obtained without culture-dependent methods (Zhou et al. 2015; Kimura 2016). This strategy allows a more detailed characterization of community patterns by providing greater depth and detection of rare and non-cultivable species (Hiergeist et al. 2015; Stefani et al. 2015; Hiraoka et al. 2016). Several studies have applied a metagenomic approach in microbes-beetle interactions, i.e. bacterial communities of Dendroctonus ponderosae (Adams et al. 2013); fungal communities associated with bark beetles (Miller et al. 2016) and fungal communities in mycangia of ambrosia beetles (Kostovcik et al. 2015).
The aim of this study was to characterize the fungal community associated with M. mutatus and different host trees through a metagenomic approach. To our knowledge, metagenomic techniques have not been previously used in investigations of forest pests from Argentina, including Platypodinae species. The aims of this research were to: 1) characterize the fungal communities associated to M. mutatus; 2) compare the culture-independent approach of the present study to a previous culture-based study (Ceriani-Nakamurakare et al. 2016), in terms of data recovery related to the composition of fungal community; and to 3) test the geographic and host-tree specificity of the assemblage of fungal communities. This research involved two host trees and three locations. In one of these locations, the fungal community has been previously analyzed by culture-dependent methods (Ceriani-Nakamurakare et al. 2016).
2 Material and methods
Fungal samples were obtained across three locations in Argentina (Fig. 1), in the phytogeographical region corresponding to the Pampa´s division (Kelt and Meserve 2014), during season 2014/2015:
(AP) Morse, Province of Buenos Aires (34°45´S, 60°53´W) at 80 m a.s.l. The plant material investigated included P. deltoides, a clone Australian I29/60. Two M. mutatus attacked plants were selected from a 12-year-old commercial poplar plantation. The plantation had a density of approximately 425 trees ha–1 (square of plantation 4 × 4 m), average diameter at breast height (DBH) 68.5 cm. The annual average rainfall ranges from 850 to 1050 mm; the average 10-year temperature shows July is the coldest month with 6.9 °C and January is the warmest month with 25 °C, with extreme temperatures that reach 41 °C in summer and –2 °C in winter (data from the National Weather Service of Argentina).
(BP) Bragado, Province of Buenos Aires (35°10´S, 60°17´W) at 68 m a.s.l. The plant material investigated included P. deltoides, a clone Stoneville 66. Two M. mutatus attacked plants were selected from a 10-year-old commercial poplar plantation. The plantation had a density of approximately 1100 trees ha–1 (square of plantation 3 × 3 m), average DBH 23.2 cm. The annual average rainfall ranges from 800 to 1000 mm; the average 10-year temperature shows July is the coldest month with 7 °C and January is the warmest month with 23.5 °C, with extreme temperatures that reach 45 °C in summer and –6 °C in winter (data from the National Weather Service of Argentina).
(CC) Concordia, Province of Entre Rios (31°25´S, 58°04´W) at 31 m a.s.l. The plant material included C. cunninghamiana (Australian beefwood), a species broadly used for shelter belts that provide wind protection for crops and animals (commonly used in citrus plantations in Argentina), and for riverbank stabilization. Therefore, M. mutatus attacked windbreak plants (average DBH 33.5 cm) were selected from a commercial citrus plantation. The annual average rainfall ranges from 786 to 2193 mm; the average 10-year temperature shows July is the coldest month with 12.5 °C and January is the warmest month with 25.3 °C, with extreme temperatures that reach 41.4 °C in summer and –5.1 °C in winter (data from the National Weather Service of Argentina).
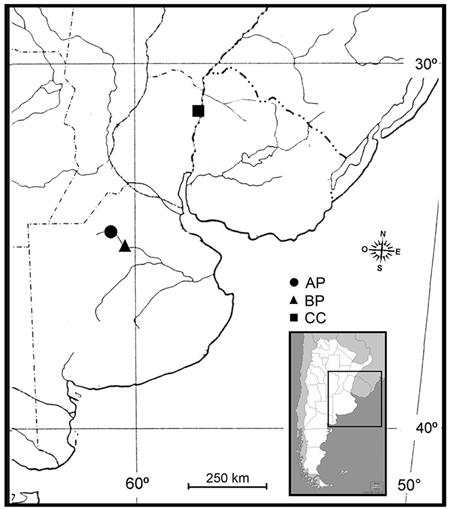
Fig. 1. Sampling locations of fungal associates to Megaplatypus mutatus in Argentina. AP: Morse, Province of Buenos Aires- Populus deltoides; BP: Bragado, Province of Buenos Aires- Populus deltoides; CC: Concordia, Province of Entre Rios- Casuarina cunninghamiana.
Active galleries were recognized according to Funes et al. (2011). Each attacked tree was cut into an individual log which contained a single active gallery; its maintenance and fragmentation was conducted according to Belhoucine et al. (2011). From each gallery, 45 representative points were randomly selected and collected with a sterile needle from the gallery surface, and pooled in 50 µL of MicroBead Solution (UltraCleanTM Microbial DNA Isolation Kit, MOBIO Laboratories MOBIO) (Table 1). The gallery samples were obtained in sterile conditions, and all harvesting instruments used were sterilized before and after each usage to prevent contamination.
| Table 1. Location, Host plants and sample collection of fungi associated to Megaplatypus mutatus galleries in Argentina. Locations are named as City and Province, respectively. | ||||||
| Sample ID | Location | Host plant | No. of trees | No. of galleries | Metagenomic samples | Collection points |
| AP | A. Morse Buenos Aires | Populus deltoides | 2 | 2 | 2 | 90 |
| BP | B. Bragado Buenos Aires | Populus deltoides | 2 | 2 | 2 | 90 |
| CC | C. Concordia Entre Ríos | Casuarina cunninghamiana | 2 | 2 | 2 | 90 |
Total DNA was extracted from collection tubes (the pooled fungal samples), using an UltraClean TM Microbial DNA Isolation Kit (MoBio Laboratories Inc., Solana Beach, USA) with the additional step for alternative lysis, in order to increase yields, following the manufacturer’s instructions. The quantity and quality of the DNA was assessed using the Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). In order to document the presence of fungal DNA in each sample, the Internal Transcribed Spacer region of the nuclear DNA (ITS) was amplified using the universal primers ITS1 and ITS4 (White et al. 1990), with the following PCR conditions: initial denaturation at 94 °C for 3 min, 40 cycles of denaturation at 94 °C for 45 s, annealing at 56 °C for 45 s, extension at 72 °C for 1 min and final extension at 72 °C for 7 min. Reactions were performed in 50.0 ml volumes containing 10 mM PCR buffer supplied by the manufacturer, 1.5 mM MgCl2, 1 mM of each dNTP, 10 mM of each primer, 1 U Recombinant GoTaq DNA polymerase (Promega Corp.) and 2 μl fungal genomic DNA. Amplification was visualized by electrophoresis of the PCR products on a 1% agarose gel stained with Gel RedTM (Biotium).
Community metagenomes were subsequently generated from the extracted DNA. Amplicon libraries were sequenced using 454-FLX-Titanium chemistry at INDEAR, Rosario, Argentina. Samples were PCR-amplified using ITS1F-ITS4 nuclear ITS rDNA fungal “barcode” to amplify the internal transcribed spacer region 2 (White et al. 1990; Schoch et al. 2012; Nilsson et al. 2015). The standard Roche A-adaptor and a unique 10 bp MID (Multiplex IDentifier) tag for each sample (within collection) were attached to the primers. Pyrosequencing was carried out following the standard procedures suggested by the manufacturer, and raw data was processed using QIIME 1.8.0 (Caporaso et al. 2010). OTUs were clustered at 97% sequence similarity. For each OTU the most abundant read was designated as the representative sequence. Taxonomic assignment was made using the naïve Bayesian classifier (minimum confidence set to 0.8) against the UNITE database. Sequence data were deposited in the GenBank/NCBI as BioProject ID PRJNA453387, PRJNA453390 and PRJNA453397.
To compare the fungal diversity among the samples, the sampling depth was normalized to those of the sample with the smallest number of reads by random removal of sequencing reads. Accumulation curves of observed OTUs of each sample and diversity indexes were calculated. In order to quantify differences in the fungal communities among samples (beta diversity), a matrix of pairwise Bray–Curtis dissimilarities was generated and a Principal Coordinate Analysis (PCoA) was made to visualize sample similarities with the effect of host plant. Pyrosequencing data and all statistical analysis were conducted using QIIME 1.8.0 (Caporaso et al. 2010) following the methods according to Orgiazzi et al. (2013), unless otherwise stated. Operational Taxonomic Units (OTUs) that were supported by fewer than 4 sequences were removed (Miller et al. 2016), while singletons (clusters containing only one read) were discarded as they are indistinguishable from sequencing errors (Kunin et al. 2010). OTUs were clustered at 97% sequence similarity. For each OTU the most abundant read was designated as the representative sequence (six samples-dataset). To refine the assignments previously made (vs UNITE database), MegaBLAST searches against the NCBI database and the MycoBank database were conducted (locations-dataset) (Altschul et al. 1990; Robert et al. 2013). In all cases the consensus sequences obtained from 97% similarity analyses were compared with the datasets obtained from Ceriani-Nakamurakare et al. (2016). The reference sequences used were published by expert taxonomists and/or deposited in culture collections (if available, sequences of type strains were preferred). A statistical analysis was performed in order to compare the fungal richness in each dataset (R Software Core Development Team 2016, version 3.3.2), employing a Generalized Linear Mixed Models (GLMM) fit by maximum likelihood with Poisson distribution (Breslow and Clayton 1993), using the lme4 package 1.1-12 (Bates et al. 2015). Trees were used as a random variable in the GLMM analyses. Assumptions were assessed. The best-fitting model was selected by using AIC. The package “Multcomp” was used performing post hoc Tukey’s all-pairwise comparisons among the levels of the factor Site. Alpha and beta diversity of fungal communities in each location was estimated by ACE, CHAO1, Shannon, Simpson and Sorensen indexes (EstimateS version 9.1, Colwell 2013).
3 Results
The mycobiota associated with galleries of M. mutatus was characterized, and the specificity of fungal assemblages was evaluated amongst three locations in two different host plants from a total of 270 collection points, where each gallery had 45 random points pooled for pyrosequencing samples. The output of pyrosequence analyses achieved a total of 102.830 reads. After the second quality-filtering 42.965 (41.78%) sequences were removed, leaving 59.865 (58.22%) sequences for further analysis. The average read count per sample was 625 with 474–831 average reads as minimum and maximum, respectively. Additional statistics for raw sequence data is provided (Suppl. file S2).
The assembled sequences represented a total of 149 OTUs (97%-similarity) grouped into 14 fungal taxa. The relative abundance of the fungal taxa and evenness of communities are shown in Suppl. file S3. Accumulation curves and diversity indexes were generated on the six samples-dataset (Suppl. file S4 and S5, respectively). Accumulation curves of fungal communities failed to reach the asymptote, indicating that diversity estimates would increase with deeper sequencing; however, a gradual tendency to reach the plateau is present. Pyrosequence data were analyzed by GLMM and results indicated that the OTUs richness at BP site was significantly higher than for the other two sampling locations (Suppl. file S6).
The Phylum Ascomycota represented the fungal diversity across all samples with 81.2% of total reads, while the Phylum Basidiomycota achieved 18.4% of total reads and unassigned OTUs obtained 0.4%. A closer analysis showed that, within these Phyla, the predominant taxa were: Saccharomycetales (38.7 %), Hypocreales (20.6%), Xylariales (11.6%), Microascales (0.7%) and Agaricales (14.2%). To ascertain how different fungal communities were associated to host type, a principal coordinate analysis was performed (Fig. 2). Axes 1 and 2 explained 67% of the variability observed between sampled host plants. The PCoA indicated that the samples from the same site were closely related; however, the samples from the same host remained separated.
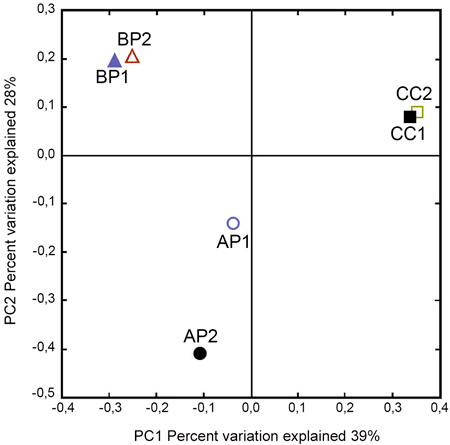
Fig. 2. Principal Coordinate Analysis plots of fungal communities according to the host plant order associated to Megaplatypus mutatus galleries. PCoA based on Bray–Curtis dissimilarities. Sampled locations in Argentina, AP: Morse, Buenos Aires- Populus deltoides; BP: Bragado, Buenos Aires- Populus deltoides; CC: Concordia, Entre Rios- Casuarina cunninghamiana.
The refined taxonomic assignments (locations-dataset) with relative frequency per location are shown in Table 2. A set of biodiversity indexes was computed (Table 3). Several taxa were highlighted either because they were locally abundant and/or present in all the surveyed locations. As shown in Table 2, C. insectalens and Fusarium sp. 1 achieved 37.6% of the total relative frequency per taxa and were the only two species present in all host plants and locations, with a frequency range of 2.4–91.4% and 0.01–7.9% per location, respectively.
| Table 2. Taxonomic assignments of associated fungi to Megaplatypus mutatus galleries in three locations. Each column represents a different location and each row a taxonomic assignment. Sampled locations in Argentina, AP: Morse, Buenos Aires- Populus deltoides; BP: Bragado, Buenos Aires- Populus deltoides; CC: Concordia, Entre Rios- Casuarina cunninghamiana. | ||||
| Total relative frequency per taxa (%) | Taxonomic assignments | Relative frequency per location (%) | ||
| AP | BP | CC | ||
| 33.71 | Candida insectalens | 2.38 | 3.99 | 91.46 |
| 15.29 | Paracremonium sp. | 0.01 | 41.77 | 0 |
| 12.78 | Phaeoisaria clematidis | 0.01 | 34.92 | 0 |
| 12.22 | Coprinellus radians | 42.24 | 0 | 0.01 |
| 6.34 | Dipodascacea | 21.93 | 0.01 | 0 |
| 4.92 | Chaetomium sp. | 17.04 | 0 | 0 |
| 3.88 | Fusarium sp. 1 | 0.01 | 3.05 | 7.98 |
| 3.45 | Coprinellus micaceus | 0 | 9.44 | 0 |
| 2.73 | Dekkera sp. | 9.44 | 0 | 0 |
| 1.98 | Fusarium sp. 2 | 0.1 | 5.35 | 0 |
| 1.08 | Paecilomyces sp. | 3.76 | 0 | 0 |
| 0.70 | Graphium sp. | 2.43 | 0 | 0 |
| 0.52 | Fusarium sp. 3 | 0.01 | 1.42 | 0 |
| 0.16 | Arthrobotrys sp. | 0.54 | 0 | 0 |
| 0.07 | Kodamaea sp. | 0 | 0 | 0.2 |
| 0.05 | Fusarium oxysporum species complex | 0 | 0 | 0.14 |
| 0.04 | Clonostachys rosea | 0 | 0 | 0.12 |
| 0.03 | Candida sp. 1 | 0.1 | 0 | 0 |
| 0.03 | Fellomyces sp. | 0 | 0.01 | 0.07 |
| 0.02 | Phialemonium dimorphosporum | 0 | 0.04 | 0.02 |
| 100 | 100 | 100 | 100 | |
| Table 3. Diversity and richness indexes from associated fungi to Megaplatypus mutatus galleries. Calculated from locations-dataset. Diversity indexes show the highest values for CC sample. The most similar set of samples was AP-BP. Sampled locations in Argentina were, AP: Morse, Buenos Aires- Populus deltoides; BP: Bragado, Buenos Aires- Populus deltoides; CC: Concordia, Entre Rios- Casuarina cunninghamiana. | ||||||||
| Sample ID | ACE | Chao1 | Shannon | Simpson | 1° Sample | 2° Sample | Shared Taxa | Sorensen |
| AP | 11.52 | 10.99 | 1.07 | 2.65 | AP | BP | 7 | 0.58 |
| BP | 17.14 | 16.92 | 1.74 | 4.56 | AP | CC | 3 | 0.27 |
| CC | 20 | 20 | 2.05 | 5.59 | BP | CC | 4 | 0.44 |
Sequences assigned to the genus Paracremonium obtained 15.29% of the total, with a range between 0.01–41.8%, and the sequences assigned to Fusarium genus achieved 6.43% of the total with a range between 0.01 and 7.98%, depending on the evaluated location. Four Fusarium species were ascribed in this study. One group of OTUs was assigned to an unidentified species (here Fusarium sp. 1), being the only Fusarium species present in all geographic locations. Furthermore, during the verification of the assignments, all the OTUs ascribed to Fusarium sp. 1 were found to be closely related to Fusarium solani species complex (FSSC). The second and third groups of OTUs were assigned to an undetermined species of Fusarium (here as Fusarium sp. 2 and Fusarium sp. 3, respectively) present in Morse and Bragado. The fourth group corresponded to Fusarium oxysporum species complex (FOSC), which was only found in Concordia. Further, species such as Coprinellus micaceus, Clonostachys rosea and sequences assigned to Dekkera sp. and Paecilomyces sp., among others, were only found in one location.
It is worth mentioning that during the taxonomic assignments, specifically during MegaBLAST searches, sequences previously obtained from the culture-based methods (Ceriani-Nakamurakare et al. 2016) were found as the match sequences (86–100% MegaBLAST Identity) in the current analysis. The current taxonomic assignment, OTU(s) number and GenBank accession number of previously obtained sequences (*) are summarized here: Fusarium sp. 1, OTUs: 42-66-68-76 with KT828712/13/14/16/18/19/20/21/22/23/25* from FSSC; FOSC, OTUs: 8-21 with KT828724*; Graphium sp., OTU: 29 with KT828732/33* from G. basitruncatum; C. radians, OTUs: 7-55-72-81-139-141-143 with KT828727/KT828739*; Dipodascaceae, OTUs: 1-6-53-100 with KT828738*. Moreover, the analysis shows a similar species recovery, with 91.67% ascribed to FSSC; while FOSC, Graphium sp., Dipodascaceae and C. radians achieved 100% each, i.e. high percentage values mean that a high number of sequences from a previous culture database were found as match sequence.
This data strengthens the congruence of molecular identification by Ceriani-Nakamurakare et al. (2016) and the metagenomics data. The diversity indexes registered lower values in Morse (AP) compared to the other locations; while Bragado (BP) and Concordia (CC) had closer values. The similarity (Sørensen’s index, Table 3) between Morse and Bragado was the highest at 0.58, whereas between Bragado–Concordia and Morse–Concordia, it had values of 0.44 and 0.27, respectively. Results from the generalized linear mixed model (locations-dataset) showed no significant differences between taxa richness amongst locations (Table 3).
4 Discussion
This research involves a metagenomics approach to explore the fungal communities associated with galleries of M. mutatus and their specificity in two different host plants and three locations in Argentina. To our knowledge, this is the first approach using metagenomic methods to investigate mycobiota associated with Platypodinae species.
We found that the Ascomycota was the most abundant Phylum. Those detected more frequently were Saccharomycetales and Hypocreales in all locations. The location or the host plant species did not have any effect on their abundance. Previously, Hu et al. (2015) reported high abundance of Saccharomycetales within galleries of Dendroctonus armandi, while Scully et al. (2014) reported that gut communities of Anoplophora glabripennis were consistently dominated by Hypocreales. These results suggest that the environmental conditions present in the gallery were suitable for these taxa. One sampled P. deltoides (Morse, AP1) tree achieved the highest abundance of C. radians, a member of Phylum Basidiomycota (91.8% of relative frequency), and the lowest abundance of members of the Phylum Ascomycota (8.2%). This fungus may have been present as an endophyte or a wood-decaying species prior to the insect attack (Eslyn and Lombard 1984; Sun et al. 2011). The results obtained during this study complement the previous data reported by Ceriani-Nakamurakare et al. (2016) representing the first comprehensive analyses on fungal communities associated to M. mutatus. Taking into account that these associations may indirectly affect bark beetle population dynamics (Popa et al. 2012), the information here analyzed improves our skills to develop new or underexplored control strategies.
Principal Coordinates Analyses show a separation between fungal communities within the same host (P. deltoides and C. cunninghamiana). These data suggested that the host identity species and anatomic characteristics are not crucial factors determining the specificity in fungal communities. In the same way, the results obtained by Sorensen index and by GLMM (significant differences for BP and no significant differences from locations-dataset) support the conclusion arise from PCoA. Taking into account that Bragado and Morse share climatic conditions and biological biotic factors (e.g. genetic inputs), it is plausible to assume that the differences observed in terms of fungal community could be related to the contrasting forestry management, which might influence the micro-environmental conditions, and/or others factors previously reported with impact on the structure of fungal assemblages (Martínez‐García et al. 2015; Schimann et al. 2016; Pérez‐Izquierdo et al. 2017).
These results have an impact in the context of biological invasions, taking into account that numerous exotic pest and pathogen introductions have led to significant ecological and/or economic damage (Hulme 2009; Ramsfield et al. 2016). Wood-infesting pests can negatively impact forest productivity, wood quality and forest ecosystem dynamics (Dreistadt et al. 1990; Liebhold et al. 1995; Boyd et al. 2013; Charles et al. 2014). The low specificity of fungal community could increase the risk in terms of biological invasion spread (Hulcr and Dunn 2011; Ploetz et al. 2013), since M. mutatus could increase the dissemination of fungal species transiently associated to the insect being plant pathogen associations or not.
Candida insectalens and Fusarium sp. 1 (ascribed to FSSC), appear in all geographic locations and host plants, highlighting the potential role of these species in the Fungi–M. mutatus–Host plant interaction. Candida species are widely recorded as yeasts associated with ambrosia beetles (Batra 1966; Francke-Grosmann 1967). This genus has been reported in associations with Platypus cylindrus, P. lewisi, P. koryoensis, among others (Henry 1967; Endoh et al. 2008; Yun et al. 2015). Candida insectalens was originally isolated from the gallery system of a platypodid ambrosia beetle Crossotarsus externedentatus infesting Cryptocarya latifolia and Ficus sycomorus in South Africa (Van der Walt 1972). Endoh et al. (2011) isolated a Candida species that was phylogenetically related to C. insectalens from P. quercivorus beetle galleries of several Quercus species and Castanopsis cuspidate in Japan, showing high frequencies among all sources analyzed. Those results supported the importance of C. insectalens in the interaction established by platypodids, independently of the host attacked. In our work currently under preparation, this species was registered as a gut-associate of M. mutatus (unpublished data).
The association between Fusarium sensu lato and ambrosia beetles has been reported in several previous publications (Freeman et al. 2013; Ceriani-Nakamurakare et al. 2016; Short et al. 2017). In this study, the presence of Fusarium species was detected in all locations investigated; Fusarium sp. 1 (FSSC), Fusarium sp. 2 and Fusarium oxysporum species complex (FOSC) were recorded and each one had a particular representativity among locations. Fusarium solani species complex (FSSC) was the only one to appear in all the locations, Fusarium sp. 2 was present in Morse and Bragado, and FOSC was only found in Concordia. Taking previous data from a culture dependent approach (Ceriani-Nakamurakare et al. 2016) into consideration, a higher relative frequency from this genus was expected; but 8.24% was the highest relative frequency recorded from Bragado, and the lowest was 0.11, from Morse. One possible explanation is the ability of the genus Fusarium to grow in culture medium and to surpass other fungal species by employing different methods, i.e. resource competition, release of metabolites and high rates of sporulation (Dighton et al. 2005; Chatterjee et al. 2016), which could lead to register high frequencies in cultures. The interaction between Fusarium species and several species of ambrosia beetles have been already reported (Kasson et al. 2013; Ceriani-Nakamurakare et al. 2016). However, it is difficult to elucidate their significance in the interactions, taking into account the many roles that this genus plays in the ecosystem, i.e. entomopathogenic, nutritional and symbiotic partner (Qi et al. 2011; Scully et al. 2012).
Phaeoisaria clematidis appeared with a high frequency in this study, mostly associated with samples obtained from Bragado. This fungus is a ubiquitous, cosmopolitan, soilborne species, and a well-known plant pathogen. It is very common in the tropics, frequently isolated on dead plant material (Guarro et al. 2000) and it has been associated with decaying wood (Tsui et al. 2003; Whitton et al. 2012). Other species of this genus have been reported associated with stained wood from bark beetle galleries in timber trees in New Zealand, Norway and Western Canada (Eyjolfsdottir 1990). It is possible to conclude that this high frequency responds to the wood decay process in sampled trees rather than a key association with the insect.
An unexpected result was almost complete absence of Graphium and Raffaelea species. During this study, only one species of Graphium was found in Morse (P. deltoides). However, during the sample collection for this study, synnematas corresponding to Graphium species were detected in all galleries from the three locations. Hence, the lack of representatives in metagenomic data may be due to the methodology used rather than a weak association in the interaction. Ceriani-Nakamurakare et al. (2016), through a culture-dependent approach and direct observation, recorded G. basitruncatum in the new segments of the gallery and on male exoskeleton. Additionally, species of Graphium have been reported as insect-vectored (Cruywagen et al. 2010), associated with Scolytodes unipunctatus (Hulcr et al. 2007; Kolařík et al. 2015) and Platypus cylindrus` mycangia (Bellahirech et al. 2014), supporting the hypothesis that this fungal genus could be relevant in the interactions.
On the other hand, Raffaelea species were not detected in this study, although this genus is frequently reported in ambrosia interactions (Ceriani-Nakamurakare et al. 2016; Ploetz et al. 2017). These results could be due to the known problems of using ITS markers with certain groups of ambrosial fungi (Fraedrich et al. 2008; Kostovcik et al. 2015). Ceriani-Nakamurakare et al. (2016), using a culture dependent approach, reported Raffaelea arxii and three additional Raffaelea species.
The results obtained through the culture-independent approach in this study and the previous culture-dependent study (Ceriani-Nakamurakare et al. 2016), were used to generate a match sequence percentage between the culture study database and the metagenomic database. This analysis indicates that the current results complement the data obtained by culturing methods. Metagenomics is a powerful tool for the characterization of fungal species diversity providing comparative results on many species and genera. Mismatches during PCR reaction may have resulted in some lineages of fungi being missed entirely; because of this, a multiple approach can improve the accuracy and the understanding of the fungal assemblages associated to M. mutatus, as was pointed out by Gazis and Chaverri (2010). Thus improving the accuracy and the understanding of the fungal assemblages associated to M. mutatus, as was pointed out by Hennessey et al. (1986).
5 Conclusions
Host plant and location did not show any significant effects on fungal richness. Regarding the obtained taxa, Fusarium and Candida species were present in all the studied associations. Finally, the composition of fungal communities associated to the beetle M. mutatus would not be determined by the identity of the host plants.
Acknowledgements
We thank Establecimiento ¨Ma. Dolores¨-Papel Prensa S.A and Establecimiento ¨San Jose¨-Flia. Euskadi, for providing technical facilities. Comments made by the journal editor and the anonymous reviewers have provided great help to improve the manuscript. This work was supported by the National Council for Scientific and Technological Research (Consejo Nacional de Investigaciones Científicas y Técnicas, CONICET) (PIP 0956, 2016–2018) and PICT-2015-1038 (2017–2019).
References
Adams A.S., Aylward F.O., Adams S.M., Erbilgin N., Aukema B.H., Currie C.R., Suen G., Raffa K.F. (2013). Mountain pine beetles colonizing historical and naive host trees are associated with a bacterial community highly enriched in genes contributing to terpene metabolism. Applied and Environmental Microbiology 79: 3468–3475. https://doi.org/10.1128/aem.00068-13.
Alfaro R.I., Humble L.M., Gonzalez P., Villaverde R., Allegro G. (2007). The threat of the ambrosia beetle Megaplatypus mutatus (Chapuis) (=Platypus mutatus Chapuis) to world poplar resources. Forestry 80: 471–479. https://doi.org/10.1093/forestry/cpm029.
Allegro G., Della Beffa G. (2001). Un nuovo problema entomologico per la pioppicoltura italiana: Platypus mutatus Chapuis (Coleoptera, Platypodidae). [A new entomological problem for poplar growing in Italy: Platypus mutatus Chapuis (Coleoptera, Platypodidae).] Sherwood Foreste ed alberi oggi 66: 31–34.
Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. (1990). Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. https://doi.org/10.1006/jmbi.1990.9999.
Bascialli M., Giménez R., Etiennot A., Toscani H. (1996). Manejo de la población de Platypus sulcatus Chap. durante tres años en la región delta del Paraná, mediante control químico. [Management of the population of Platypus sulcatus Chap. for three years in the delta region of Paraná, through chemical control]. Investigaciones Agrícolas, Sistema de Recursos Forestales 5: 129–140.
Bates D., Mächler M., Bolker B., Walker S. (2015). Fitting linear mixed-effects models using lme4. Journal of Statistical Software 67: 1–48. https://doi.org/10.18637/jss.v067.i01.
Batra L.R. (1963). Ecology of ambrosia fungi and their dissemination by beetles. Transactions of the Kansas Academy of Science 66: 213–236. https://doi.org/10.2307/3626562.
Batra L.R. (1966). Ambrosia fungi: extent of specificity to ambrosia beetles. Science 153: 193–195. https://doi.org/10.1126/science.153.3732.193.
Belhoucine L., Bouhraoua R.T., Meijer M., Houbraken J., Harrak M.J., Samson R.A., Equihua-Martinez A., Pujade-Villar J. (2011). Mycobiota associated with Platypus cylindrus (Coleoptera: Curculionidae, Platypodidae) in cork oak stands of north west Algeria, Africa. African Journal of Microbiology Research 5: 4411–4423. https://doi.org/10.5897/ajmr11.614.
Bellahirech A., Inácio M.L., Bonifácio L., Nóbrega F., Sousa E., Ben Jamâa M. (2014). Comparison of fungi associated with Platypus cylindrus F. (Coleoptera: Platypodidae) in Tunisian and Portuguese cork oak stands. IOBC/wprs Bulletin 101: 149–156.
Bobrowsky P.T. (2013). Encyclopedia of natural hazards. Springer, New York. 1135 p. https://doi.org/10.1007/978-1-4020-4399-4.
Boyd I., Freer-Smith P., Gilligan C., Godfray H. (2013). The consequence of tree pests and diseases for ecosystem services. Science 342: 1235773. https://doi.org/10.1126/science.1235773.
Breslow N.E., Clayton D.G. (1993). Approximate inference in generalized linear mixed models. Journal of American Statistical Association 88: 9–25. https://doi.org/10.1080/01621459.1993.10594284.
Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Buschman F.D., Costello E.K. (2010). QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7: 335–336. https://doi.org/10.1038/nmeth.f.303.
Castrillo L.A., Griggs M.H., Vandenberg J.D. (2016). Competition between biological control fungi and fungal symbionts of ambrosia beetles Xylosandrus crassiusculus and X. germanus (Coleoptera: Curculionidae): Mycelial interactions and impact on beetle brood production. Biological Control 103: 138–146. https://doi.org/10.1016/j.biocontrol.2016.09.005.
Ceriani-Nakamurakare E., Slodowicz M., Gonzalez-Audino P., Dolinko A., Carmarán C. (2016). Mycobiota associated with the ambrosia beetle Megaplatypus mutatus: threat to poplar plantations. Forestry 89: 191–200. https://doi.org/10.1093/forestry/cpw001.
Charles J., Nef L., Allegro G., Collins C.M., Delplanque A., Gímenez R., Höglund S., Jiafu H., Larsson S., Luo Y., Parra P., Singh A.P., Volney W.J.A., Augustin S. (2014). Insect and other pests of poplars and willows. In: Isebrands J.G., Richardon J. (eds.). Poplars and willows: trees for society and the environment. CABI, Oxford. p. 459–526. https://doi.org/10.1079/9781780641089.0459.
Chatterjee S., Kuang Y., Splivallo R., Chatterjee P., Karlovsky P. (2016). Interactions among filamentous fungi Aspergillus niger, Fusarium verticillioides and Clonostachys rosea: fungal biomass, diversity of secreted metabolites and fumonisin production. BMC Microbiology 16: 83. https://doi.org/10.1186/s12866-016-0698-3.
Colwell R.K. (2013). EstimateS, Version 9.1: statistical estimation of species richness and shared species from samples (Software and User’s Guide). Freeware for Windows and Mac OS. http://viceroy.uconn.edu/estimates/index.html.
Cruywagen E.M., De Beer Z.W., Roux J., Wingfield M.J. (2010). Three new Graphium species from baobab trees in South Africa and Madagascar. Persoonia-Molecular Phylogeny and Evolution of Fungi 25: 61–71. https://doi.org/10.3767/003158510x550368.
Dighton J., White Jr J.F., White J., Oudemans P. (2005). The fungal community: its organization and role in the ecosystem, 3rd ed. CRC Press, New York. 960 p. https://doi.org/10.1201/9781420027891.
Dreistadt S.H., Dahlsten D.L., Frankie G.W. (1990). Urban forests and insect ecology. BioScience 40: 192–198. https://doi.org/10.2307/1311364.
Endoh R., Suzuki M., Benno Y., Futai K. (2008). Candida kashinagacola sp. nov., C. pseudovanderkliftii sp. nov. and C. vanderkliftii sp. nov., three new yeasts from ambrosia beetle-associated sources. Antonie van Leeuwenhoek 94: 389–402. https://doi.org/10.1007/s10482-008-9256-9.
Endoh R., Suzuki M., Okada G., Takeuchi Y., Futai K. (2011). Fungus symbionts colonizing the galleries of the ambrosia beetle Platypus quercivorus. Microbial Ecology 62: 106–120. https://doi.org/10.1007/s00248-011-9838-3.
Eslyn W.E., Lombard F.F. (1984). Fungi associated with decayed wood in stored willow and cottonwood logs. Mycologia 76: 548–550. https://doi.org/10.2307/3793339.
Eyjolfsdottir G.G. (1990). Fungi isolated from stained wood associated with bark beetle galleries in timber trees in New Zealand, Norway and Western Canada. Ph.D. thesis, University of Manitoba.
Fraedrich S., Harrington T., Rabaglia R., Ulyshen M.D., Mayfield III A.E., Hanula J.L., Eickwort J.M., Miller D.R. (2008). A fungal symbiont of the redbay ambrosia beetle causes a lethal wilt in redbay and other Lauraceae in the southeastern United States. Plant Disease 92: 215–224. https://doi.org/10.1094/pdis-92-2-0215.
Francke-Grosmann H. (1956). Hautdrüsen als träger der pilzsymbiose bei ambrosiakäfern. [Skin glands as a carrier of fungal symbiosis in ambrosia beetles]. Zeitschrift für Morphologie und Öekologie der Tiere 45: 275–308.
Francke-Grosmann H. (1967). Ectosymbiosis in wood-inhabiting insects. Symbiosis 2: 141–205. https://doi.org/10.1016/b978-1-4832-2758-0.50010-2.
Freeman S., Sharon M., Maymon M., Mendel Z., Protasov A., Aoki T., Eskalen A., O’Donell K. (2013). Fusarium euwallaceae sp. nov. a symbiotic fungus of Euwallacea sp., an invasive ambrosia beetle in Israel and California. Mycologia 105: 1595–1606. https://doi.org/10.3852/13-066.
Funes H., Griffo R., Zerba E., Gonzalez-Audino P. (2011). Mating disruption of the ambrosia beetle Megaplatypus mutatus in poplar and hazelnut plantations using reservoir systems for pheromones. Entomologia Experimentalis et Applicata 139: 226–234. https://doi.org/10.1111/j.1570-7458.2011.01126.x.
Gazis R., Chaverri P. (2010). Diversity of fungal endophytes in leaves and stems of wild rubber trees (Hevea brasiliensis) in Peru. Fungal Ecology 3(3): 240–254. https://doi.org/10.1016/j.funeco.2009.12.001.
Gebhardt H., Begerow D., Oberwinkler F. (2004). Identification of the ambrosia fungus of Xyleborus monographus and X. dryographus (Coleoptera: Curculionidae, Scolytinae). Mycological Progress 3: 95–102. https://doi.org/10.1007/s11557-006-0080-1.
Giménez R.A., Etiennot A.E. (2007). Rango de hospederos de Platypus mutatus (Chapuis, 1865) (Coleoptera: Platypodidae). [Host range of Platypus mutatus (Chapuis, 1865) (Coleoptera: Platypodidae)]. Entomotropica 18: 89–94.
González-Audino P., Gatti P., Zerba E. (2011). Traslucent pheromone traps increase trapping efficiency of ambrosia beetle Megaplatypus mutatus. Crop Protection 30: 745–747. https://doi.org/10.1016/j.cropro.2011.02.008.
Guarro J., Vieira L.A., De Freitas D., Gené J., Zaror L., Hofling-Lima A.L., Fischman O., Zorat-Yu C., Figueras M.J. (2000). Phaeoisaria clematidis as a cause of keratomycosis. Journal of Clinical Microbiology 38: 2434–2437.
Hennessey T.C., Dougherty P.M., Kossuth S.V., Johnson J.D. (1986). Stress physiology and forest productivity: proceedings of the physiology working group technical session. Martinus Nijhoff Publishers, Dordrecht. 239 p. https://doi.org/10.1007/978-94-009-4424-4.
Henry S.M. (1967). Symbiosis: associations of invertebrates, birds, ruminants, and other biota. Academic Press, New York & London. 462 p.
Hiergeist A., Gläsner J., Reischl U., Gessner A. (2015). Analyses of intestinal microbiota: culture versus sequencing. ILAR Journal 56: 228–240. https://doi.org/10.1093/ilar/ilv017.
Hiraoka S., Yang C., Iwasaki W. (2016). Metagenomics and bioinformatics in microbial ecology: current status and beyond. Microbes Environment 31: 204–212. https://doi.org/10.1093/ilar/ilv017.
Hu X., Li M., Chen H. (2015). Community structure of gut fungi during different developmental stages of the Chinese white pine beetle (Dendroctonus armandi). Scientific Reports 5: 8411. https://doi.org/10.1038/srep08411.
Hulcr J., Dunn R.R. (2011). The sudden emergence of pathogenicity in insect–fungus symbioses threatens naive forest ecosystems. Proceedings of the Royal Society of London: Biological Sciences 278(1720): 2866–2873. https://doi.org/10.1098/rspb.2011.1130.
Hulcr J., Stelinski L.L. (2016). The ambrosia symbiosis: from evolutionary ecology to practical management. Annual Review Entomology 62: 285–303. https://doi.org/10.1146/annurev-ento-031616-035105.
Hulcr J., Kolarik M., Kirkendall L.R. (2007). A new record of fungus-beetle symbiosis in Scolytodes bark beetles (Scolytinae, Curculionidae, Coleoptera). Symbiosis 43: 151.
Hulme P.E. (2009). Trade, transport and trouble: managing invasive species pathways in an era of globalization. Journal of Applied Ecology 46: 10–18. https://doi.org/10.1111/j.1365-2664.2008.01600.x.
Kasson M.T., O’Donnell K., Rooney A.P., Sink S., Ploetz R.C., Ploetz J.N., Konkol J.L., Carrillo D., Freeman S., Mendel Z., Smith J.A., Black A.W., Hulcr J., Bateman C., Stefkova K., Campbell P.R., Geering A.D.W., Dann E.K., Eskalen A., Mohotti K., Short D.P.G., Aoki T., Fenstermacher K.A., Davis D.D., Geiser D.M. (2013). An inordinate fondness for Fusarium: phylogenetic diversity of fusaria cultivated by ambrosia beetles in the genus Euwallacea on avocado and other plant hosts. Fungal Genetics and Biology 56: 147–157. https://doi.org/10.1016/j.fgb.2013.04.004.
Kelt D.A., Meserve P.L. (2014). Status and challenges for conservation of small mammal assemblages in South America. Biological Reviews Cambridge Philosophical Society 89: 705–722. https://doi.org/10.1111/brv.12080.
Kimura N. (2006). Metagenomics: access to unculturable microbes in the environment. Microbes Environment 21: 201–215. https://doi.org/10.1264/jsme2.21.201.
Kolařík M., Hulcr J., Kirkendall L.R. (2015). New species of Geosmithia and Graphium associated with ambrosia beetles in Costa Rica. Czech Mycology 67: 29–35.
Kostovcik M., Bateman C.C., Kolarik M., Stelinski L.L., Jordal B.H., Hulcr J. (2015). The ambrosia symbiosis is specific in some species and promiscuous in others: evidence from community pyrosequencing. The ISME Journal 9: 126–138. https://doi.org/10.1038/ismej.2014.115.
Kunin V., Engelbrektson A., Ochman H., Hugenholtz P. (2010). Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environmental Microbiology 12: 118–123. https://doi.org/10.1111/j.1462-2920.2009.02051.x.
Liebhold A.M., MacDonald W.L., Bergdahl D., Mastro V.C. (1995). Invasion by exotic forest pests: a threat to forest ecosystems. Forest Science 41: a0001-z0001.
Martínez‐García L.B., Richardson S.J., Tylianakis J.M., Peltzer D.A., Dickie I.A. (2015). Host identity is a dominant driver of mycorrhizal fungal community composition during ecosystem development. New Phytologist 205: 1565–1576. https://doi.org/10.1111/nph.13226.
Miller K.E., Hopkins K., Inward D.J., Vogler A.P. (2016). Metabarcoding of fungal communities associated with bark beetles. Ecology and Evolution 6: 1590–600. https://doi.org/10.1002/ece3.1925.
Nilsson R.H., Tedersoo L., Ryberg M., Kristiansson E., Hartmann M., Unterseher M., Porter T.M., Bengtsson-Palme J., Walker D.M., de Sousa F., Gamper H.A., Larsson E., Larsson K-H., Kõljalg U., Edgar R.C., Abarenkov K. (2015). A comprehensive, automatically updated fungal ITS sequence dataset for reference-based chimera control in environmental sequencing efforts. Microbes Environment 30: 145–150. https://doi.org/10.1264/jsme2.me14121.
Orgiazzi A., Bianciotto V., Bonfante P., Daghino S., Ghignon S., Lazzari A., Lumini E., Mello A., Napoli C., Perotto S., Vizzini A., Bagella S., Murat C., Girlanda M. (2013). 454 pyrosequencing analysis of fungal assemblages from geographically distant, disparate soils reveals spatial patterning and a core mycobiome. Diversity 5: 73–98. https://doi.org/10.3390/d5010073.
Pérez‐Izquierdo L., Zabal‐Aguirre M., Flores‐Rentería D., González‐Martínez S.C., Buée M., Rincón A. (2017). Functional outcomes of fungal community shifts driven by tree genotype and spatial‐temporal factors in Mediterranean pine forests. Environmental Microbiology 19: 1639–1652. https://doi.org/10.1111/1462-2920.13690.
Ploetz R.C., Hulcr J., Wingfield M.J., de Beer Z.W. (2013). Destructive tree diseases associated with ambrosia and bark beetles: black swan events in tree pathology? Plant Disease 97: 856–872. https://doi.org/10.1094/pdis-01-13-0056-fe.
Ploetz R.C., Konkol J.L., Narvaez T., Duncan R.E., Saucedo R.J., Campbell A., Mantilla J., Carrillo D., Kendra P.E. (2017). Presence and prevalence of Raffaelea lauricola, cause of laurel wilt, in different species of ambrosia beetle in Florida, USA. Journal of Economic Entomology 110: 347–354. https://doi.org/10.1093/jee/tow292.
Popa V., Déziel E., Lavallée R., Bauce E., Guertin C. (2012). The complex symbiotic relationships of bark beetles with microorganisms: a potential practical approach for biological control in forestry. Pest management science 68(7): 963–975. https://doi.org/10.1002/ps.3307.
Qi H., Wang J., Endoh R., Takeuchi Y., Tarno H., Futai K. (2011). Pathogenicity of microorganisms isolated from the oak platypodid, Platypus quercivorus (Murayama) (Coleoptera: Platypodidae). Applied Entomology and Zoology 46: 201–210. https://doi.org/10.1007/s13355-011-0032-3.
Ramsfield T., Bentz B., Faccoli M., Jactel H., Brockerhoff E. (2016). Forest health in a changing world: effects of globalization and climate change on forest insect and pathogen impacts. Forestry 89: 245–252. https://doi.org/10.1093/forestry/cpw018.
Robert V., Vu D., Amor A.B.H., van de Wiele N., Brouwer C., Bernard J., Szoke S., Dridi A., Triki M., Ben Daoud S., Chouchen O., Vaas L., de Cock A., Stalpers J.A., Stalpers D., Verkley G., Groenewald M., Borges dos Santos F., Stegehuis G., Li W., Wu L., Zhang R., Ma J., Zhou M., Pérez Gorjón S., Eurwilaichitr L., Ingsriswang S., Hansen K., Schoch C., Robbertse B., Irinyi L., Meyer W., Cardinali G., Hawksworth D.L., Taylor J.W., Crous P.W. (2013). MycoBank gearing up for new horizons. IMA fungus 4: 371–379. https://doi.org/10.5598/imafungus.2013.04.02.16.
Schimann H., Bach C., Lengelle J., Louisanna E., Barantal S., Murat C., Buée M. (2016). Diversity and structure of fungal communities in neotropical rainforest soils: the effect of host recurrence. Microbial Ecology 73: 310–320. https://doi.org/10.1007/s00248-016-0839-0.
Schoch C.L., Seifert K.A., Huhndorf S., Robert V., Spouge J.L., Levesque C.A., Chen W., Fungal Barcoding Consortium (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America 109(16): 6241–6246. https://doi.org/10.1073/pnas.1117018109.
Scully E.D., Hoover K., Carlson J., Tien M., Geib S.M. (2012). Proteomic analysis of Fusarium solani isolated from the Asian longhorned beetle, Anoplophora glabripennis. PloS one 7: e32990. https://doi.org/10.1371/journal.pone.0032990.
Scully E.D., Geib S.M., Carlson J.E., Tien M., McKenna D., Hoover K. (2014). Functional genomics and microbiome profiling of the Asian longhorned beetle (Anoplophora glabripennis) reveal insights into the digestive physiology and nutritional ecology of wood feeding beetles. BMC Genomics 15: 1096. https://doi.org/10.1186/1471-2164-15-1096.
Short D.P., O’Donnell K., Stajich J.E., Hulcr J., Kijimoto T., Berger M.C., Macias A.M., Spahr E.J., Bateman C.C., Eskalen A., Lynch S.C., Cognato A.I., Cooperband M.F., Kasson M.T. (2017). PCR multiplexes discriminate Fusarium symbionts of invasive Euwallacea ambrosia beetles that inflict damage on numerous tree species throughout the United States. Plant Disease 101: 233–240. https://doi.org/10.1094/pdis-07-16-1046-re.
Stefani F.O., Bell T.H., Marchand C., de la Providencia I.E., El Yassimi A., St-Arnaud M., Hijri M. (2015). Culture-dependent and-independent methods capture different microbial community fractions in hydrocarbon-contaminated soils. PloS one 10: e0128272. https://doi.org/10.1371/journal.pone.0128272.
Sun X., Guo L.-D., Hyde K. (2011). Community composition of endophytic fungi in Acer truncatum and their role in decomposition. Fungal Diversity 47: 85–95. https://doi.org/10.1007/s13225-010-0086-5.
Tremblay E., Espinosa B., Mancini D., Caprio G. (2000). Un coleottero proveniente dal Sudamerica minaccia i pioppi. [A beetle from South America threatens poplars]. Informatore Agrario 56: 89–90.
Tsui C.K., Hyde K.D., Fukushima K. (2003). Fungi on submerged wood in the Koito River, Japan. Mycoscience 44: 55–59. https://doi.org/10.1007/s10267-002-0083-y.
Van der Walt J. (1972). The yeast genus Ambrosiozyma gen. nov. (Ascomycetes). Mycopathologia et Mycologia Applicata 46: 305–315. https://doi.org/10.1007/bf02052126.
White T.J., Bruns T., Lee S., Taylor J.W. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J. (eds.). PCR protocols: a guide to methods and applications. Academic Press, San Diego, California. p. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1.
Whitton S.R., McKenzie E.H., Hyde K.D. (2012). The current understanding of fungi associated with Pandanaceae. Fungal Diversity Research Series 21: 1–428. https://doi.org/10.1007/978-94-007-4447-9_1.
Yun Y.H., Suh D.Y., Yoo H.D., Oh M.H., Kim S.H. (2015). Yeast associated with the ambrosia beetle, Platypus koryoensis, the pest of oak trees in Korea. Mycobiology 43: 458–466. https://doi.org/10.5941/myco.2015.43.4.458.
Zhou J., He Z., Yang Y., Deng Y., Tringe S.G., Alvarez-Cohen L. (2015). High-throughput metagenomic technologies for complex microbial community analysis: open and closed formats. MBio 6: e02288-02214. https://doi.org/10.1128/mbio.02288-14.
Total of 76 references.

