Abiotic stresses induced physiological, biochemical, and molecular changes in Betula platyphylla: a review
Ritonga F. N., Ngatia J. N., Song R. X., Farooq U., Somadona S., Lestari A. T., Chen S. (2021). Abiotic stresses induced physiological, biochemical, and molecular changes in Betula platyphylla: a review. Silva Fennica vol. 55 no. 3 article id 10516. https://doi.org/10.14214/sf.10516
Highlights
- Abiotic stress influence Betula platyphylla growth, development, and yield production by impairing physiological, biochemical, and molecular functions
- Overexpression or RNAi line of transcription factors enhance the abiotic stress tolerance of B. platyphylla
- MYB and AP2/ERF are the most frequently transcription factor family that has been explored over the last two decades in B. platyphylla under abiotic stress.
Abstract
Abiotic stress is one of the major factors in reducing plant growth, development, and yield production by interfering with various physiological, biochemical, and molecular functions. In particular, abiotic stress such as salt, low temperature, heat, drought, UV-radiation, elevated CO2, ozone, and heavy metals stress is the most frequent study in Betula platyphylla Sukaczev. Betula platyphylla is one of the most valuable tree species in East Asia facing abiotic stress during its life cycle. Using transgenic plants is a powerful tool to increase the B. platyphylla abiotic stress tolerance. Generally, abiotic stress reduces leaves water content, plant height, fresh and dry weight, and enhances shed leaves as well. In the physiological aspect, salt, heavy metal, and osmotic stress disturbs seed germination, stomatal conductance, chlorophyll content, and photosynthesis. In the biochemical aspect, salt, drought, cold, heat, osmotic, UV-B radiation, and heavy metal stress increases the ROS production of B. platyphylla cells, resulting in the enhancement of enzymatic antioxidant (SOD and POD) and non-enzymatic antioxidant (proline and AsA) to reduce the ROS accumulation. Meanwhile, B. platyphylla upregulates various genes, as well as proteins to participate in abiotic stress tolerance. Based on recent studies, several transcription factors contribute to increasing abiotic stress tolerance in B. platyphylla, including BplMYB46, BpMYB102, BpERF13, BpERF2, BpHOX2, BpHMG6, BpHSP9, BpUVR8, BpBZR1, BplERD15, and BpNACs. These transcription factors bind to different cis-acting elements to upregulate abiotic stress-related genes, resulting in the enhancement of salt, drought, cold, heat, osmotic, UV-B radiation, and heavy metal tolerance. These genes along with phytohormones mitigate the abiotic stress. This review also highlights the candidate genes from another Betulacea family member that might be contributing to increasing B. platyphylla abiotic stress tolerance.
Keywords
antioxidant;
cis-acting elements;
gene;
ROS production;
transcription factor
- Ritonga, State Key Laboratory of Tree Genetics and Breeding, Forestry College, Northeast Forestry University, Harbin 150040, China E-mail ritongafaujiah@ymail.com
- Ngatia, College of Wildlife and Protected Areas, Northeast Forestry University, Harbin 150040, China E-mail jacob.ngatia3@gmail.com
- Song, State Key Laboratory of Tree Genetics and Breeding, Forestry College, Northeast Forestry University, Harbin 150040, China E-mail 13359850710@163.com
- Farooq, College of Life Science, Northeast Forestry University, Harbin 150040, China E-mail uf@nn.ch
- Somadona, College of Agriculture, Riau University, Pekanbaru 28293, Indonesia E-mail sonia_hut@yahoo.co.id
- Lestari, Forestry Major, College of Agriculture, Mataram University, Mataram 83125, Indonesia E-mail atlestari@unram.ac.id
-
Chen,
State Key Laboratory of Tree Genetics and Breeding, Forestry College, Northeast Forestry University, Harbin 150040, China
E-mail
chensu@nefu.edu.cn

Received 23 January 2021 Accepted 14 June 2021 Published 18 June 2021
Views 132313
Available at https://doi.org/10.14214/sf.10516 | Download PDF

Supplementary Files
1 Introduction
White birch (Betula platyphylla Sukaczev) is treated lately as a synonym of Betula pendula subsp. mandshurica (WCSP 2013; Shaw et al. 2015) is one of the most economically important tree species in boreal areas of America, Eupore, and East Asia (Ashburner and McAllister 2013; Lv et al. 2019) including Northern China (Guo et al. 2017a, 2017b), and Northern Japan (Hoshika et al. 2013b). It is a deciduous and pioneer woody plant (Guo et al. 2017a) that has heterophyllous leaves (Hoshika et al. 2013b) and is a cold-tolerant species (Chen et al. 2019). This species belongs to the Betulaceae family (IPNI 1911; WCSP 2013) and shares the same genus as B. pendula Roth (Lemmetyinen et al. 2008; Gang et al. 2019a, 2019b; Wang et al. 2019a), B. luminifera H.J.P. Winkl. (Pan et al. 2017b), and B. ermanii Cham. (Muraoka and Koizumi 2005; Wang et al. 2018). Betula platyphylla has been widely used for health due to its betulin, betulinic acid, phenolic, and oleanolic acid content (Keinänen et al. 1999; Fan et al. 2014; Razieh et al. 2018; Ma et al. 2019; Yin et al. 2020), and also in papermaking, architecture, and furniture due to special characteristics of its wood, including hard, elastic, and uniform structure as well (Sun et al. 2012; Zhao et al. 2019).
Forest is an important aspect to improve the global greening and ecological environment. Illegal logging and converting natural forest area into agricultural, forest industry area, and oil palm plantation (Ritonga et al. 2018), biotic stress (Iason et al. 2018; Li et al. 2020b; Perea et al. 2020), and abiotic stress (Cocozza et al. 2009; Estravis-Barcala et al. 2020; Zhou et al. 2020) are the important factors that affect forest performance and wood productivity. However, comparing with crops, studies of abiotic stress tolerance of forest trees, and research on resistance breeding are still in their infancy (Niinemets 2010). This is mainly because woody plant breeding is very slow due to its large size, long generation cycle, as well as need a long time to test tree economic value (Lemmetyinen et al. 2008). Abiotic stresses are increasing, and only a few tree species can grow normally in such conditions. It is known that abiotic stresses, including cold, drought, osmotic, high salinity, UV-radiation, and heavy metals stress are hostile to plant growth and development, causing yield reduction (Hussain 2019) and worldwide economic losses (He et al. 2018). As such, abiotic stress tolerance research in tree species is critical (Zhang et al. 2020). To date, several studies about physiological, biochemical, and molecular analysis of B. platyphylla under abiotic stress have been done including transcriptome analysis of B. platyphylla under low-temperature stress (Yan et al. 2020), analysis of the B. platyphylla auxin response factor (BpARF) expression in B. platyphylla under drought stress (Li et al. 2020a), and physiological and molecular analysis of B. platyphylla under salt stress (Mijiti et al. 2017), in spite of the fact that they still remain a need for further studies.
Plants defend themselves against abiotic stresses by scavenging reactive oxygen species (ROS) (Deng et al. 2018; Dreyer and Dietz 2018), stress acclimations such as cold acclimation, drought acclimation, and salinity acclimation (Kargiotidou et al. 2010; Banik et al. 2016; Kamanga et al. 2020), enzymatic and non-enzymatic antioxidant enhancement (Bankaji et al. 2019), and so on. Besides, transcription factor (Lotfi et al. 2019) has an important role in abiotic stress, enhancing gene expression associated with abiotic stress response at the transcription level (Zhang et al. 2020). The TF family has numerous members; unfortunately, the functions of the majority of that TF are lacking, especially in B. platyphylla. Some physiological measurements such as electrolyte leakage (EL), chlorophyll content, photosynthesis, and biochemical measurements, including malondialdehyde (MDA), antioxidant enzymes, and proline content was used to confirm the molecular analysis (Almeida et al. 2013; Chai et al. 2017; Lopez-Delacalle et al. 2020). Previous studies indicated that intrinsic physiological and biochemical mechanisms contributed to plant tolerance under abiotic stress, such as ozone (O3), water stress, and drought stress conditions (Šircelj et al. 2007; Pellegrini et al. 2019). Although the physiological, biochemical, and molecular mechanisms of some plants involved in the stress response regulation, growth, and development have been emphasized, the data is still limited and indistinct, and the regulatory network of gene expressions needs to be further elucidated. In this paper, we perform review aimed at elucidating and characterizing the mechanisms of B. platyphylla under abiotic stresses, as well as shortlisting them to identify a common pathway in physiological, biochemical and molecular aspects. This is the first literature review of B. platyphylla under abiotic stress.
2 General effects of abiotic stresses on Betula platyphylla
Most studies on the response of B. platyphylla under abiotic stress have been largely performed in Asia. The interest of East Asian countries towards this topic could be largely influenced by the fact that B. platyphylla is one of the most valuable Eastern Asia tree species. The environmental condition is one of the biggest factors influencing plant growth and production (Zhang et al. 2020). Recently, many studies about environmental stress in plants have increased providing new insights into physiological, biochemical, and molecular aspects (Shao et al. 2018; Riikonen et al. 2020; Ritonga and Chen 2020). The dominance of psychological, biochemical, and molecular aspects in B. platyphylla studies under abiotic stress was shown in Fig. 1.
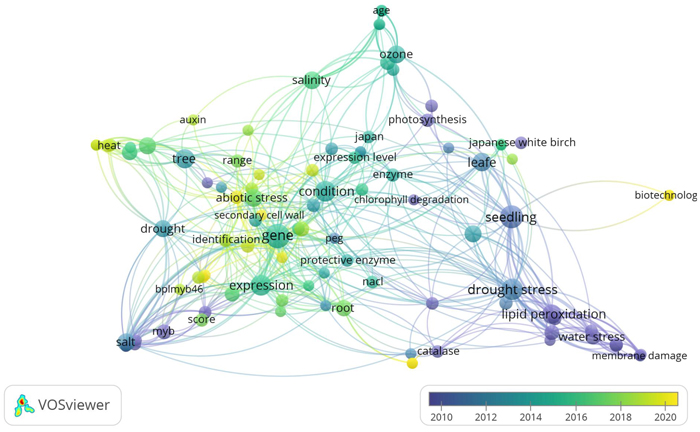
Fig. 1. Term co-occurrence map of physiological, biochemical, and molecular aspects in Betula platyphylla studies under abiotic stress using VOSviewer software (https://www.vosviewer.com/). VOSviewer analyzed text mining and bibliometric of scientific papers by observing the outputs of term (keyword) co-occurrence analysis. It is obvious that drought stress, heat, salinity, ozone, and water stress as well, are the most dominant abiotic stress, but the other key terms such as physiological and biochemical parameters belong to the other terms.
Fig. 1 showed that abiotic stresses such as salinity, drought, O3, heat, water stress, and osmotic stress were the most occurrence abiotic stress studies on B. platyphylla. For physiological and biochemical analysis, lipid peroxidation, protective enzymes, and photosynthesis were the most occurrence, while the expression of genes, BplMYB46, and MYB TF family were the most occurrence in molecular aspects. Considering the close connections among all aspects, this section provides the impact of abiotic stress on various morphological, phenotype, physiological, and biochemical parameters.
2.1 Morphological and phenotype adaptation of Betula platyphylla under abiotic stress
The plants face combinations of stresses during tree ontogeny (Niinemets 2010). Each plant has its intraspecific variation in abiotic stress tolerance (Hannus et al. 2021). Furthermore, the effects of abiotic stress on tree performance are difficult to analyze (Niinemets 2010). Several morphological and phenotypic parameters are normally measured to analyze the effect of abiotic stress on plants, such as roots (diameter, root length, and root weight) (Arif et al. 2020), leaf (number of stems and leaf area) (Malinowska et al. 2020), and flower (anther development, pollen variability, filament elongation, and ovule) (Su et al. 2013). A previous study has revealed that B. platyphylla has tissue-specific characteristics under different abiotic stress (Tan et al. 2020). Betula platyphylla responds to abiotic stress by having some morphological and anatomical adaptation. Wen et al. (2019) revealed that an injured phenotype was acquired in drought-treated B. platyphylla than that in normal conditions. The leaves were drooped followed by the reduction of leaves’ water content (Wen et al. 2019). Under O3 stress, the number of attached leaves at the B. platyphylla seedling terminal shoots initially increased, and then several leaves had an early decline that showed by the enhanced of shed leaves (Hoshika et al. 2013b). Under normal conditions, there was no difference in morphological and phenotype such as height, growth, root size, root weight, and fresh weight among B. platyphylla homeobox-leucine zipper 2 gene (BpHOX2) overexpression (OE) lines, repressed-expression (RE), and WT plants. However, BpHOX2 OE lines showed the green leaves under osmotic stress, while RE and WT plants showed yellow leaves. Similar to leaves, the height of all plants was different under osmotic stress. OE lines had the tallest followed by WT, and then the RE lines. The fresh weight, roots size, and root weight of OE lines were also higher compared with those of the RE lines and WT plants (Tan et al. 2020). Under salt and osmotic stress, the plant height, fresh and dry weight of Ethylene Responsive Factor 11 (BpERF11) RNA-interference (RNAi) lines were the highest, followed by the WT and OE lines, respectively. The RNAi lines also had lower water loss compared with WT plants and OE lines (Zhang et al. 2016).
2.2 Abiotic stress affects secondary metabolite on Betula platyphylla
Plants, as renewable resources, provide phytochemicals that are known as secondary metabolites to tolerate stress (Christie et al. 1994; Mahajan et al. 2020). Secondary metabolites function in the plant adaptation encountering environmental stress conditions (Ali and Abbas 2003). Plant secondary metabolites can be categorized into four major classes, namely terpenoid, phenolic, alkaloid, and sulfur compound (Guerriero et al. 2018). The secondary metabolite accumulation occurred when plants were subjected to stresses (biotic and abiotic) through signal molecules and various elicitors as well (Akula and Ravishankar 2011). Secondary metabolite products such as flavonoid and betulin were elevated under abiotic stresses in B. platyhylla (Popov et al. 2017; Yin et al. 2020). A flavonoid is a natural substance group along with variable phenolic structures. Flavonoid is found in bark, leaves, even roots of the Betulaceae family including B. pendula, B. pubescens Ehrh., B. papyrifera Marshall, as well as B. platyphylla (Keinänen and Julkunen-Tiitto 1998; Germanò et al. 2012; Riikonen et al. 2020). This natural product is well known for its beneficial effects on pharmacological, medicinal, nutraceutical, and cosmetic application due to its ability to defend against pathogens, tumor inhibition, anti-inflammatory, anti-HIV, anti-oxidative, anti-mutagenic, anti-carcinogenic, pneumonia, nephritis, chronic bronchitis, and choloplania coupled with its capacity to modulate the function of the key cellular enzyme (Germanò et al. 2012; Oh et al. 2012; Eom et al. 2016; Panche et al. 2016; Yin et al. 2020). The effect of abiotic stress on the secondary metabolite of B. platyphylla was also pointed out by (Riikonen et al. 2020). Combined O3 stress and ambient CO2 reallocate carbon through terpenoids biosynthesis to phenolic acids, resulting in increased phenolic compounds. Interestingly, CO2 excess and ambient O3 increased the content of phenolic compounds by elevating the phenolic acid content (Riikonen et al. 2020).
Similar to phenolics, the secondary metabolism of B. platyphylla also produces triterpenoids which have an important role in pharmacological activities, including anti-HIV, anti-AIDS, anti-cancer, tumor inhibition, arthritis, and defense against pathogens (Ju et al. 2004; Huh et al. 2011; Yin et al. 2020). The increase in the accumulation of triterpenes (betulin and betulinic acid) in B. pendula was significantly higher after elicitation by chitosan, methyl jasmonate (MeJA), pectin, and yeast extract than that of other elicitors (Razieh et al. 2018). According to (Yin et al. 2020), triterpene synthesis in B. platyphylla was affected by the expression of two birch oxidosqualene cyclase (OSC) genes, namely BpCAS and Bpβ-AS. Phytohormones such as MeJA, gibberellin (GA3), abscisic acid (ABA), and ethylene along with mechanical damage induced BpCAS and Bpβ-AS. The inhibition of Bpβ-AS positively regulates the synthesis of betulinic acid which is a naturally occurring pentacyclic triterpenoid. Whereas the BpCAS RNAi line can significantly promote the conversion of 2,3-oxidosqualene to the downstream products of betulinic and oleanolic acid. Furthermore, betulin production was also influenced by the endophytic fungus Phomopsis (Phomopsis spp.). It was revealed that Phomopsis spp. can stimulate the secondary metabolite biosynthesis in B. platyphylla (Fan et al. 2014). Nowakowska et al. (2020) also claimed that Phytophthora cactorum (Lebert and Cohn) J. Schröt and Armillaria gallica Marxm. and Romagn., soil-borne pathogens could increase the production of phenols and triterpenes, suggesting plant systemic acquired resistance (SAR) was activated. Besides, the hydrogen sulfide (H2S) donor sodium hydrosulfide (NaHS) could enhance the B. platyphylla betulin production. NaHS also promoted the gene expression of squalene, farnesyl pyrophosphate, and lupeol synthase related to betulin synthesis in B. platyphylla transgenic plants by RNAi method (Ma et al. 2019).
2.3 Photosynthesis changes in Betula platyphylla under abiotic stresses
One of the most important physicochemical processes in plants is photosynthesis. However, photosynthesis is highly sensitive to abiotic stresses. Abiotic stress inhibits photosynthesis to cope with osmotic changes in the plant (Hussain 2019). Abiotic stress leads to a reduction in the photosynthesis process by decreasing chlorophyll content (Khan et al. 2020), stomatal conductance, and net photosynthetic rate (Feng et al. 2009), also increasing ROS and suffering membrane integrity (Dubey et al. 2021). Due to suffered membrane integrity, the plant photosynthesis process was inhibited, as well as intercellular CO2, electron transport, stomatal conductance, transpiration, photochemical and non-photochemical, maximum quantum yield, and operating efficiency of PSII (Mijiti et al. 2017; Dubey et al. 2021). Abiotic stress significantly reduced the photosynthesis process in B. platyphylla by reducing chlorophyll content and/or increasing the chlorophyll degradation (Guo et al. 2017b; Lv et al. 2020a). It was found that Brassinazole-Resistant 1 (BpBZR1) expression contributed to maintaining photosynthetic intensity and reducing chlorophyll degradation under salt stress (Lv et al. 2020a). Rather, myeloblastosis 46 (BplMYB46) OE lines increased the photosynthesis compared to control due to BplMYB46 appears to be involved in controlling stomatal aperture to minimize water loss. Besides, BplMYB46 increased secondary cell wall thickness and lignin deposition, and also reduces cell death under salt and osmotic stress (Guo et al. 2017b).
Contrastingly, BpERF11 OE lines showed an increase in water loss under salt and osmotic stress. Stomatal aperture is closely related to the increased water loss under salt and osmotic stress in B. platyphylla. Interestingly, BpERF11 RNAi lines showed a reduced stomatal aperture compared with control, illustrating that BpERF11 is a negative regulator of stomatal aperture to reduce water loss (Zhang et al. 2016). Zhang et al. (2016) also revealed that the chlorophyll contents were also lower in OE lines, and the RNAi lines displayed the highest levels. Furthermore, BpMYB61 functions in controlling the stomatal aperture. Zhang et al. (2016) analyzed the expression of myeloblastosis 61 (BpMYB61) in transgenic B. platyphylla, and found that BpMYB61 was downregulated in BpERF11 OE lines, but upregulated in the RNAi lines, illustrating that BpERF11 downregulated the BpMYB61 expression.
Generally, the photosynthesis and growth rate of B. platyphylla did not decrease under ambient O3 exposure (Hoshika et al. 2013a). Meanwhile, according to (Hoshika et al. 2013b), O3 stress did not reduce photosynthesis in early leaves of B. platyphylla, illustrating that early leaves were less susceptible under O3 stress than late leave. It was assumed that early leaves can avoid O3-induced stress due to the reduction in stomatal conductance (Gs) and enhancement of N content. Lower Gs and greater N content were assumed to contribute to the O3 acclimation process. The findings demonstrated that early and late leaves have different functional roles in O3 stress tolerance in B. platyphylla (Hoshika et al. 2013b). Furthermore, Kinose et al. (2014) found that lower optimal temperature for stomatal opening and rapid leaf maturation enhances the cumulative stomatal O3 uptake (COU) of B. platyphylla under O3 stress.
The limited N treatment caused the reduction of non-regulated non-photochemical quenching rate (_JNO) resulted in the enhanced loss of absorbing light, indicated by the increase of leaf chlorophyll (JChl) in B. platyphylla. This result was assumed as protection of B. platyphylla to adapt to further abiotic stresses (Kitao et al. 2019). Under elevated CO2 and low N treatment, the accumulation of sugar and starch causing electron transport rate (ETR) reduction and non-photochemical quenching (NqP) enhancement. NqP equilibrates the energy dissipation reduction through ETR and still faces a higher risk of photoinhibition from excessive excitation energy in PSII, resulted in a reduction in photochemical quenching (qP) in B. platyphylla seedlings. This impaired photoprotection capacity caused B. platyphylla to become more vulnerable to photoinhibition in the event of additional abiotic stresses including cold, salt, heat, or drought stress (Kitao et al. 2005). Therefore, abiotic stress in B. platyphylla leads to the reduction of photosynthesis by enhancing the stomatal aperture and water loss, and chlorophyll degradation as well.
2.4 Phytohormones response of Betula platyphylla to abiotic stress
One of the first plant responses to tolerate abiotic stress is to regulate photosynthesis for survival which is related to the changes of phytohormone pathways. Phytohormones are abscisic acid (ABA), Exogenous 6-benzyl amino purine (6BA), auxins, brassinosteroids, salicylic acid (SA), ethylene, gibberellic acid (GA), cytokinin, and jasmonic acid (JA). Phytohormones have numerous functions in the plant such as growth and development and mitigating and impairing abiotic stress effects (Ciarkowska et al. 2016; An et al. 2019). Endogenous and exogenous phytohormones rescue plants from abiotic stress by regulating enzymes (Ciarkowska et al. 2016) and increasing antioxidant activities (Torun et al. 2020). In current years, crosstalk of hormone with the response of B. platyphylla to abiotic stress is starting to emerge. GA3 and 6BA hormone mitigated the effect of salt stress on B. platyphylla seed germination. By contrast, exogenous melatonin and naphthalene acetic acid (NAA) treatment exacerbated the effect of salt stress on seed germination (Li et al. 2019). The expression of BpNAC012 OE lines in B. platyphylla, a gene from NAM (no apical meristem), ATAF 1/2 (Arabidopsis transcription activator factor 1/2), and CUC2 (cup-shaped cotyledon) TF family, was induced by ABA and abiotic stress (Hu et al. 2019). A similar result was also found in B. halophila Ching ex P.C. Li (An et al. 2019). In B. platyphylla, the germination rate of ultraviolet radiation 8 genes (BpUVR8) transgenic lines and control plants decreased significantly after ABA supplemented. Furthermore, compared to control plants, BpUVR8 OE lines seed germination was remarkably inhibited. It was assumed that the BpUVR8 enhances the susceptibility to ABA, demonstrating ABA regulates BpUVR8 and can inhibit the germination of seed (Li et al. 2018). The early response to dehydration 15 gene (BplERD15) expression that was induced by drought, salt, and osmotic stress, was also inhibited under ABA treatment in B. platyphylla transgenic lines (Lv et al. 2020b).
2.5 Antioxidant defense of Betula platyphylla under abiotic stress
During abiotic stress, ROS induction causes oxidative stress in plants. Plants confer facilitation to enzymatic antioxidant and non-enzymatic antioxidant machinery. Antioxidants mitigate abiotic stress to lead to abiotic stress tolerance. Generally, the antioxidant enzymes which get elevated are peroxidase (Oberschelp et al. 2020), superoxide dismutase (SOD), catalase (CAT), glutathione S-transferase (GST), and ascorbate peroxidase (APX), whereas non-enzymatic antioxidant that gets increased is proline, and ascorbic acid (AsA) (Liu et al. 2018; Lv et al. 2020a). Firstly, abiotic stress results in ROS accumulation, and SOD was enriched to lead superoxide anion (O2–) conversion to H2O2. - POD and CAT converse the toxic H2O2 in B. platyphylla cells. The H2O2 and O2– levels were significantly elevated under drought stress, and MDA as well (Wen et al. 2019). Under salt and osmotic stress, SOD and POD activities increased significantly in BpNAC012 OE lines compared to control plants. Furthermore, the SODs and PODs genes were higher in OE lines, demonstrating that BpNAC012 could induce SODs and PODs expression (Hu et al. 2019).
BplMYB46 affects ROS scavenging under salt and osmotic stress tolerance in B. platyphylla. By using 3,3-diaminobenzidine (Alisoltani et al. 2019) in situ staining, it was revealed that the main ROS species, H2O2, was reduced in OE plants. The activities of SOD and POD were also higher compared to RE lines and control plants under salt and osmotic stress. Similar to BpNAC012, BplMYB46 improves salt and osmotic tolerance by regulating the expression of SOD, POD, and P5CS genes to increase ROS scavenging and proline contents (Guo et al. 2017b). The BplMYB46 and BplMYB13 co-expressing transgenic plants displayed higher ROS scavenge abilities and increased SOD, POD, and GST activities compared to plants overexpressing BplMYB13 or BplMYB46 alone under salt and osmotic stress conditions. BplMYB46 and BplMYB13 co-overexpressing transgenic plants decreased cell death, illustrating that the interaction between BplMYB46 and BplMYB13 could enhance salt and osmotic tolerance (Wang et al. 2019b).
High-Mobility Group 6 (BpHMG6) gene enhanced POD activity, and ROS scavenging capacity to alleviate cell damage and death by salinity stress in B. platyphylla (Lei et al. 2021). The overexpression of BpBZR1 reduced H2O2 accumulation increased antioxidant enzyme activities and maintained high photosynthetic intensity by reducing chlorophyll degradation. The results demonstrated that BpBZR1 OE lines enhanced the salt tolerance in B. platyphylla (Lv et al. 2020a). Nitro Blue Tetrazolium (NBT) staining also can be used as evidence to show the accumulation of ROS under stress conditions. The ROS accumulation and MDA content of BpUVR8 OE lines increased under ABA treatment in transgenic Arabidopsis thaliana (L.) Heynh., which demonstrated that O2– and H2O2 were involved in the response of the Arabidopsis UVR8-mediated to ABA (Li et al. 2018). NBT and DAB staining were used to measure O2– and H2O2 levels in Ethylene Response Factor 13 (BpERF13) OE lines under cold stress, Heat shock protein 9 gene (BpHSP9) OE lines under heat stress, and BpHOX2 OE lines under osmotic stress. It was revealed that O2– and H2O2 were significantly reduced compared to control and RNAi line plants. It was also reported that BpERF13, BpHSP9, and BpHOX2 enhanced the ROS scavenging, SOD and POD activity, and non-enzymatic antioxidants, resulting in increased freezing, heat, and osmotic tolerance, respectively (Liu et al. 2018; Lv et al. 2019; Tan et al. 2020). Further, BpHOX2 influenced the proline biosynthesis genes, pyrroline-5-carboxylate synthase (BpP5CS1 and BpP5CS2) under osmotic stress. The transcript level of BpP5CS1 and BpP5CS2 were significantly higher in the BpHOX2 OE lines, illustrated that BpP5CS1 and BpP5CS2 were induced by BpHOX2, resulted in the enhancement of proline level in transgenic B. platyphylla under osmotic stress (Tan et al. 2020). Consistent with previous abiotic stresses, H2O2 and O2− levels were markedly increased under Cd stress in B. platyphylla control plants. Consistently, ROS accumulation was enhanced followed by MDA content. However, B. platyphylla overexpressing of Long non-coding RNAs (LncRNA28068.1 and LncRNA2705.1) had lower H2O2 and O2− levels under Cd stress, suggesting that these LncRNAs confer Cd tolerance in B. platyphylla (Wen et al. 2020). The list of antioxidant enzymes involved under abiotic stress in OE or RNAi lines was shown in Table 1.
| Table 1. The information of antioxidant enzymes that are used as potential markers of abiotic stress tolerance in Betula platyphylla. All studies were conducted in China from 2017–2020. | |||
| Antioxidant enzymes | Type of abiotic stress | Response | References |
| SOD, POD | salt and osmotic stress | SOD and POD content in BpMYB46 OE lines increased under salt and osmotic stress | (Guo et al. 2017b) |
| SOD, POD | drought stress | SOD and POD increased in BpARF1 RNAi plant under drought stress | (Li et al. 2020a) |
| SOD, POD | heat stress | SOD and POD content in BpHSP9 OE lines increased under heat stress | (Liu et al. 2018) |
| SOD, POD | cold stress | SOD and POD content in BPERF13 OE lines increased under cold stress | (Lv et al. 2019) |
| SOD, POD | osmotic stress | SOD and POD content in BpHOX2 OE lines increased under osmotic stress | (Tan et al. 2020) |
| SOD, POD, GST | salt and osmotic stress | SOD, POD, and GST content in BplMYB46 and BplMYB13 OE lines increased under salt and osmotic stress | (Wang et al. 2019b) |
| SOD, POD | salt stress | SOD and POD content increased under salt stress in Betula platyphylla | (Mijiti et al. 2017) |
| SOD: Superoxide dismutase; POD: Peroxidase; GST: Glutathione S-transferase; BplMYB46: Betula platyphylla myeloblastosis 46 gene; BpHOX2: B. platyphylla homeobox-leucine zipper 2 gene; BpHSP9: B. platyphylla heat shock protein 9 gene; BpUVR8: B. platyphylla UV resistance locus 8 gene; BpERF13: B. platyphylla ethylene response factor 13 gene; BpMYB13: B. platyphylla myeloblastosis 13 gene; BpARF1: B. platyphylla auxin response factor 1 gene. | |||
Contrary to OE lines, BpARF1 RNAi lines reduced the ROS accumulation, increased the POD, SOD, AsA, and proline contents under drought stress in B. platyphylla. These results demonstrate that BpARF1 negatively regulates drought tolerance in B. platyphylla (Li et al. 2020a). BpERF11 OE lines had lower SOD and POD activity, and higher MDA content and H2O2 and O2–accumulation compared to RNAi line of BpERF11 under salt and osmotic stress in B. platyphylla, illustrating that BpERF11 negatively regulate membrane lipid peroxidation (Zhang et al. 2016). Several exogenous applications can be used to reverse the negative effect of abiotic stress in plants such as the application of silicon (Si). Si application could reverse the negative effects of manganese (Mn) stress in Cucumis sativus L.. The activity of APX, dehydroascorbate reductase (DHAR), and glutathione reductase (GR) enhanced under Mn stress after Si application, illustrating that Si is responsible for the lower accumulation of H2O2. Si also contributed to reducing lipid peroxidation of chloroplast and GPX induced by excess Mn, demonstrating that reduction in GPX might be one of the important mechanisms of Si for inhibiting necrosis by keeping higher photosynthesis in C. sativus (Feng et al. 2009). However, there is no information about this topic in B. platyphylla.
2.6 Electrolyte leakage
EL is related to cell membrane damage-causing cell death. Several plants are sensitive to abiotic stress, withered, and then died. Despite B. platyphylla is an abiotic stress tolerance tree species, EL value was found to be increased by abiotic stress (Zhao et al. 2014; Beloiu et al. 2020). Using Evans Blue Staining and the EL rate assay, it was shown that drought stress damaged the B. platyphylla cell membrane (Wen et al. 2019). Li et al. (2020a) found that BpARF1 RNAi lines reduce the EL and water loss, while BpARF1 OE lines showed the opposite physiological changes under drought stress. Meanwhile, the BplERD15 OE line showed a lower EL rate under drought stress, suggesting that BplERD15 is an abiotic stress-responsive gene that can reduce cell death under drought stress conditions (Lv et al. 2020b). Under cold stress, Lv et al. (2019) revealed that BpERF13 OE lines showed a higher EL rate compared to WT plants under cold stress. BpHMG6 OE lines showed lower cell damage and death (Lei et al. 2021). The ability of BpHSP9 to increase ROS scavenging, antioxidant enzymes, and non-enzymatic antioxidant activities leads to diminishing O2– and H2O2 accumulation, as well as EL reduction under heat stress in B. platyphylla. However, the reversed results were found in BpHSP9 RNAi lines (Liu et al. 2018). Contrastingly, BpERF11 RNAi lines showed lower EL and cell death salt and osmotic stress in B. platyphylla (Zhang et al. 2016). BpHOX2 OE lines had higher osmotic tolerance by protecting the cell from death compared to RE and WT plants. To confirm the EL value in OE lines, cell death was determined by using Evans Blue Staining analysis, where the cell death was higher in RE lines than those in OE lines and WT (Tan et al. 2020).
A similar result was also revealed in the BplMYB46 and BplMYB13 co-expressing lines. The less intense blue staining was found in the BplMYB46 and BplMYB13 co-expressing lines than that in BplMYB13 or BplMYB46 alone or WT plants under salt and osmotic stress. Furthermore, EL rate was lower in the BplMYB46 and BplMYB13 co-expressing transgenic plants compared to BplMYB13 or BplMYB46 alone or WT plants. These results demonstrated that BplMYB46 and BplMYB13 co-overexpressing transgenic plants could reduce the ROS accumulation and mortality, illustrating that the interaction between BplMYB46 and BplMYB13 could increase salt and osmotic tolerance in B. platyphylla (Wang et al. 2019b). When B. platyphylla was exposed to Cd stress, the EL value result demonstrated that the cell membrane was substantially damaged under excess Cd conditions. However, the LncRNA28068.1 and LncRNA2705.1 OE lines showed lower EL compared to WT and other LncRNAs such as LncRNA 11415.1 and LncRNA30505.2, illustrating that LncRNA28068.1 and LncRNA2705.1 could regulate Cd tolerance in B. platyphylla (Wen et al. 2020).
3 The molecular mechanism involved in Betula platyphylla under abiotic stress
Recently, the combination of genomics and transcriptomics is used to obtain a new insight and deep understanding of the molecular response of B. platyphylla to abiotic stress. Gene expression profiles are effective tools that can be used to investigate plant stress tolerance mechanisms. Many genes involved in abiotic stress respond to B. platyphylla to produce mRNA, as well as protein that plays a vital role in abiotic stress tolerance. Genetically transformed B. platyphylla plants were obtained from leaf discs co-cultivation with Agrobacterium tumefaciens (Smith and Townsend) Conn. The produced calli in the presence of kanamycin confirmed the successful transformation (Mohri et al. 1997). The list of B. platyphylla gene expressions under abiotic stress was displayed in Supplementary file S1.
3.1 Salt and osmotic stress
It is well known that salinity is one of the most important factors in reducing plant growth and yield all over the earth. Moreover, salinity results in osmotic stress, which causes disturbances at the metabolic level. Forest has an important role in improving the global greening and ecological environment; however, studies about salt tolerance trees, germplasm selection, and resistance breeding are limited compared to crop plants. Salinized land area is increasing, and few tree species has ability to cope and grow in salt areas, resulting in salt tolerance tree research become more serious (Zhang et al. 2020).
The study of TFs is an important part of functional genomics research. TFs is a key role in various biological processes under abiotic stress by binding to cis-acting elements to control their target gene expressions. Moreover, the gene structure is different among the different groups resulted in divergent functions. But, the distributions of most proteins in the same subfamily exhibited identical motif, which demonstrated functional similarities among the same subfamily in the same group (Guo et al. 2017a). Among the TFs related to B. platyphylla abiotic stress, the ethylene-responsive factor (ERF) and myeloblastosis oncogene (MYB) families were the most abundant, especially to salt and osmotic tolerance (Wen et al. 2019). Studies about the regulatory specificity of ERF and MYB TFs and their interactions with other factors which regulate salt and osmotic tolerance in B. platyphylla will be explained in this section. BpERF11 is a nuclear protein, which could specifically bind to GCC boxes and DRE motifs under salt and severe osmotic stress. BpERF11 OE lines showed the inhibited AtMYB61 homologous gene expression, resulting in stomatal aperture enhancement, which increased the transpiration rate. Furthermore, it was found that BpERF11 downregulated the Delta-1-pyrroline-5-carboxylate synthase (P5CSs), SOD, and POD gene expressions, while (Proline Dehydrogenase 1 (PRODH) and Delta-1-pyrroline-5-carboxylate dehydrogenase (P5CDH) were upregulated, resulting in the increased ROS accumulation and reduced proline levels. Besides, BpERF11 also significantly prevents the Late embryogenesis abundant (LEA) and dehydrin gene expression, which is involved in abiotic stress tolerance. These results demonstrated that BpERF11 is a negative regulator of B. platyphylla under salt and osmotic stress (Zhang et al. 2016).
BplMYB46 contributed to salt and osmotic tolerance in B. platyphylla. It was confirmed by using Chromatin immunoprecipitation (ChIP) assays that BplMYB46 binds to E-box, GT-box, and TC-box elements to activate b-glucuronidase (GUS) expression (Guo et al. 2018). BplMYB46 had a contribution in enhancing the expression level of proline biosynthesis genes (P5CS1 and P5CS2). Guo et al. (2017b) revealed that P5CS1 and P5CS2 expression were higher in BplMYB46 OE lines. Interestingly, proline degradation genes (P5CDH and ProDH) were highly expressed in RE lines than that in OE lines under salt and osmotic stress, demonstrating that BplMYB46 regulated salt and osmotic tolerance in B. platyphylla (Guo et al. 2017b, 2018). Similar to Guo et al. (2017b), the BpHOX2 induced the transcript levels of proline biosynthesis genes (BpP5CS1 and BpP5CS2) in BpHOX2 OE lines. BpP5CS1 and BpP5CS2 expression were significantly higher in the BpHOX2 OE lines compared to BpHOX2 RE lines and WT, indicating that BpHOX2 induces the proline content due to the up-regulation of proline biosynthesis genes under osmotic stress in B. platyphylla (Tan et al. 2020). Meanwhile, BpHOX2 could regulate the antioxidant genes such as SOD and POD genes. The expression levels of antioxidant genes were significantly increased in the BpHOX2 OE lines compared to RE lines. Furthermore, using a combination of Chip-seq and RNA-seq analysis, it was found that to regulate gene expression, BpHOX2 could bind to four cis‐acting elements, including Mybp binding box “CCWACC”, dehydration responsive element “RCCGAC”, and two novel cis‐acting elements namely “AAGAAG” and “TACGTG” which is termed as HBS1 and HBS2, respectively. The findings demonstrated that BpHOX2 increase osmotic tolerance in B. platyphylla by binding to different cis‐acting elements to regulate gene expression (Tan et al. 2020).
The NAC TFs play vital roles in plant biological processes, xylem development, and stress responses. BpNAC012 is a transcriptional activator in B. platyphylla under salt and osmotic stress. In order to enhance salt and osmotic tolerance in B. platyphylla, the core sequence CGT[G/A] was activated by BpNAC012 to induce P5CSs, SODs, and PODs expression. Moreover, BpNAC012 specifically binds to the SNBE site which is located in the promoters of secondary wall-associated genes. BpNACs were highly found in the N terminal NAC domain (A-E regions) and the C-terminal domain (variable transcriptional regulation region) (Guo et al. 2017a). BpNAC012 also participated in secondary wall thickening of stem fibers. BpNAC012 RNAi lines dramatically reduce secondary wall thickening of stem fibers. Rather, the expression of secondary wall-associated downstream genes was activated by BpNAC012 in BpNAC012 OE lines by directly binding to the secondary wall NAC-binding element sites. Consequently, resulting in ectopic secondary wall deposition in the stem epidermis (Hu et al. 2019). Furthermore, RT-qPCR analysis revealed that BpNAC012 could increase the lignin biosynthetic gene expressions, as well as lignin accumulation. The elevated lignin biosynthesis is a precarious factor in plant adaptation and tolerance during salt stress conditions (Chun et al. 2019). The results illustrated that BpNAC012 has distinct roles in salt stress responses and secondary cell wall biosynthesis as well (Hu et al. 2019).
Using sequence analysis, it was found that the BpBZR1 belongs to the BES1 subfamily. BpBZR1 is a transcriptional activator which contains 113–234 amino acid (aa) fragment that is required for activating transcription. BZR multiple sequence alignment of B. platyphylla and A. thaliana showed that there is a high similarity in the N conserved region; however, a certain difference in the C conserved region was found, suggesting the distinct function of BZR genes in B. platyphylla and A. thaliana. Furthermore, the BpBZR1 protein sequence is found to be highly conserved and similar to the BZR proteins of other plants such as Ziziphus jujube Mill, Juglans regia L., and Vitis vinifera L.. More importantly, BpBZR1 has an N-terminal, C-terminal, serine-rich phosphorylation sites and a bHLH structure. Interestingly, the BpBZR1 expression level was down-regulated after salt stress, but overexpression of BpBZR1 increased salt tolerance in B. platyphylla. Lv et al. (2020a) assumed that BpBZR1 might associate with several stress-related cis-elements to arrange a series of downstream genes expression. Consequently, higher ROS scavenger and photosynthetic strength, and lower lipid peroxidation and chlorophyll degradation (Lv et al. 2020a). The expression levels of genes are varying under abiotic stress. A similar result was revealed by Lei et al. (2021). It was found that BpHMGs in leaves, roots, and stems were varying under salt and osmotic stress. In stems, most BpHMGs genes were mainly upregulated, while most BpHMGs genes were downregulated in roots under osmotic stress. Under salt stress, most BpHMGs expressions both in roots and stems were significantly down-regulated (Lei et al. 2021).
| Table 2. The summary of cis-acting element of Betula platyphylla related to abiotic stress genes. | ||||
| No | Gene name | cis-acting element | Target genes | Reference |
| 1 | BplMYB46 | - TC-BOX ((T(G/A)TCG(C/G))) - GT-BOX (A(G/T)T(A/C)GT(T/G)C) - E-BOX ((CA(A/T/C)(A/G/C)TG) | - SOD gene - POD gene, PAL gene | (Guo et al. 2018) |
| 2 | BplMYB46 | - MYBCORE: CAGTTA - AC-box: ACCACCT | - SOD, POD, and GST | (Guo et al. 2017b; Wang et al. 2019b) |
| 3 | BpHSP | - TATA box - heat shock element (5’-AAAAAATTTC-3’) | - N.A. | (Liu et al. 2018) |
| 4 | BpARF | - 2010 bp upstream of the 5-UTRs of the BpARF genes, including: 1. abscisic acid responses 2. anaerobic induction 3. auxin responses 4. cell cycle regulation 5. gibberellin responses 6. light responses 7. methyl jasmonate (MeJA) responses 8. MYBHv1 binding 9. MYB binding in response to drought 10. salicylic acid responses 11. defense and stress responses 12. meristem expression 13. MYB binding in response to light and low-temperature responses | - N.A. | (Li et al. 2020a) |
| 5 | BpUVR8 | - The jasmonic acid methyl ester response element (CGTCA-motif) - Abscisic acid response element (ABRE) - Auxin response element (CATATG-motif) - Drought-induced response elements (MBS) - Low-temperature, heat stress - Anaerobic response elements | - N.A. | (Li et al. 2018) |
| 6 | BpERF13 | - LTRECOREATCOR15 (CCGAC):CAGGCGTCGG - MYBCORE (CNGTTR): TCAACAGGAT | - POD6 and POD8 gene - SOD1, SOD3, POD6, POD8, CBF3 and CBF4 | (Lv et al. 2019) |
| 7 | BpHOX2 | - Dehydration responsive element “RCCGAC” - Mybp binding box “CCWACC” - Novel cis‐acting elements with the sequences of “AAGAAG” - Novel cis‐acting elements with the sequences of “TACGTG” | - N.A. | (Tan et al. 2020) |
| 8 | BpERF11 | - GCC boxes - DRE motifs | - SODs, PODs, P5CS, P5CDH, PRODH, MYB61, DHN, and LEAs | (Zhang et al. 2016) |
| 9 | BpNAC012 | - The core sequence CGT[G/A] - The SNBE site | - P5CS1, P5CS2, SODs, PODs - Secondary wall biosynthesis genes (lignin, cellulose, xylem) and wood associated TF genes (MYB46, MYB54, MYB63, MYB85, KNAT7) | (Hu et al. 2019) |
| BplMYB46: Betula platyphylla myeloblastosis 46 gene; BpHOX2: B. platyphylla homeobox-leucine zipper 2 gene; BpHSP9: B. platyphylla heat shock protein; BpUVR8: B. platyphylla UV resistance locus 8 gene; BpERF13: B. platyphylla ethylene response factor 13 gene; BpERF11: B. platyphylla ethylene responsive factor 11 gene; BpARF1: B. platyphylla auxin response factor gene; BpNAC012: B. platyphylla No apical meristem (NAM), Arabidopsis transcription activation factor (ATAF1/2), Cup-shaped cotyledon (CUC2) 12 gene; SODs: Superoxide dismutase; PODs: Peroxidase; PALs: Phenylalanine ammonia lyase; GSTs: Glutathione S-transferase; CBFs: C-repeat binding factors; P5CS: Delta-pyrroline-5-carboxylate synthase; P5CDH: Delta-1 pyrroline-5-carboxylate dehydrogenase; PRODH: Proline dehydrogenase; MYBs: Myeloblastosis genes; DHN: dehydration gene; LEAs: Late embryogenesis abundant; KNAT7: Knotted-like homeobox of Arabidopsis thaliana 7; SNBE: Secondary wall NAC binding element; N.A.: no detailed comments. | ||||
3.2 Low-temperature stress
To identify the TFs involved in low-temperature tolerance, Yan et al. (2020) used transcriptome analysis and revealed that hundreds of TFs were essentially expressed under low-temperature stress in B. platyphylla. The low-temperature TFs family on B. platyphylla are APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF), Cysteine-2/Histidine-2 (C2H2), MYB-HB-like, WRKY, Basic helix-loop-helix (bHLH), WD or beta-transducin repeat-like (WD40-like), and GIBBERELLIC ACID INSENSITIVE (GAI), REPRESSOR OF GA1 (RGA), and SCARECROW (SCR) GRAS families. Furthermore, low-temperature stress significantly upregulated Calcium-binding EF-hand family protein, Calmodulin-like CML38, calmodulin-like CML25, calmodulin-like protein, N-acetyl-1-glutamate synthase, POD, and branched-chain amino acid transferase (Yan et al. 2020). As previously mentioned, BpERF13, an AP2/ERF TF family member, could enhance cold tolerance in B. platyphylla (Lv et al. 2019). To understand the BpERF13 mechanism, a TF-centered yeast one-hybrid (Y1H) experimental system was used. The result showed that to activate a reporter gene, BpERF13 binds to MYBCORE cis-elements and LTRECOREATCOR15. To strengthen the result, ChIP-seq and ChIP-PCR experiments were further used and revealed that BpERF13 bound to these cis-elements when present in the 5’ proximal regions of SOD1, SOD3, POD6, POD8, and C-repeat-binding factor (CBF3 and CBF4) genes. The list of the cis-acting element of B. platyphylla under abiotic stress was summarized in Table 2. Using qRT-PCR analysis, the expression levels of SOD, POD, and CBF genes of OE lines were significantly upregulated in response to cold stress compared to WT plants. These results demonstrate that BpERF13 regulates physiological processes underlying cold tolerance in B. platyphylla (Lv et al. 2019). BpNACs were differentially expressed under low temperature, as well as BpNACs associated gene co-expression networks. The promoters of eight BpNACs (BpNAC016, BpNAC024, BpNAC043, BpNAC052, BpNAC063, BpNAC075, BpNAC077, and BpNAC108) contain low-temperature response-related motifs, indicating that BpNACs may play important roles in cold resistance in B. platyphylla. Interestingly, it was found that the promoter of BpNAC041 contains cis-acting elements is not only involved in low temperature but also drought and salt stress (Chen et al. 2019). Recently, Chen et al. (2021) revealed that protein kinases can be connected to the network of gene regulatory systems which is involve many TFs and cold tolerance related genes by additional mitogen activated protein (MAP) and the presence of the MEKK1–MKK2–MPK4 cascade.
3.3 Heat stress
BpHSP gene is the only heat-related gene that is published in B. platyphylla. Using PLACE database, the cis-acting elements in promoter regions of 21 BpHSP genes were analyzed. Two important elements, including the TATA box and heat shock element (HSE, 5’-AAAAAATTTC-3’), were found in the promoters of all BpHSP. Heat shock factors (Hsfs) genes were assumed to regulate the BpHSPs expression. BpHsfs elevate the heat shock genes expression in response to various stimuli. The finding suggested that BpHsfs might regulate the BpHSPs expression in response to heat stimuli through HSE modules (Liu et al. 2018).
In B. luminifera, microRNAs (miRNAs) is important to regulate specific stress-responsive genes such as TFs and genes encoding important enzymes, which were related to modulation of attenuated plant growth and development, protein synthesis, protein folding, transport and turnover, cell wall organization, antioxidation, and defense response under heat stress. Using qRT-PCR and high-throughput sequencing revealed that the trend of expression patterns of 15 randomly selected miRNAs showed similarity, as well as the expression patterns of six target genes under heat stress, suggesting that miRNA mediated regulatory network of heat stress response in B. luminifera, and might contribute to improving heat stress resistant breeding in other woody plants include B. platyphylla (Pan et al. 2017b). The candidate genes that might be contributed to improving abiotic stress tolerance in B. platyphylla were shown in Table 3.
| Table 3. The summary of candidate genes for Betula platyphylla under abiotic stress. | ||||||||
| No | Family of TF/gene/miRNAs | Gene name | Species | Type of abiotic stress | Degree/ dose | Location | Application | References |
| 1 | MYB | - BplMYB46 | - Betula platyphylla | - Drought stress | N.A. | - nucleus | - BplMYB46 bind to MYBCORE in transgenic B. platyphylla, indicating BplMYB46 may contribute to drought stress. | (Guo et al. 2017b) |
| 2 | - AHL | - B. halophila | - Salt stress | 200 mM of NaCl for 24 h | - nucleus | - AHL gene was the most up-regulated gene in leaves under salt stress, indicating that the AHL gene might contribute in response to salt stress in B. halophila. | (Shao et al. 2018) | |
| - DHNs | - B. halophila | - Salt stress | 200 mM of NaCl for 24 h | - nucleus | - Dehydrin-1 gene was up-regulated gene in leaves after salt stress, indicating that dehydrin-1 gene might contribute to the response to salt stress in B. halophila. | (Shao et al. 2018) | ||
| 3 | miRNAs (miR395c-3p) | - B. luminifera | - Heat stress | - 0.5 h and 4 h | - N.A. | - miRNAs (miR395c-3p) were up-regulated under heat stress in B. luminifera, suggesting that miR395c-3p contribute to improving heat tolerance in other birches such as B. platyphylla. | (Pan et al. 2017a) | |
| 4 | NAC | - BpNAC5, - BpNAC6 BpNAC7, BpNAC8 - BpNAC13, BpNAC14, BpNAC15, BpNAC19, BpNAC21 | - B. platyphylla | - Drought, salt, dehydration, osmotic and ABA treatment | - N.A. | - nucleus and C-terminal | - Multiple copies of MBS, W-box and ABRE were found in the promoters of BpNACs, implying that these genes might play important roles in stress responses. | (Guo et al. 2017a) |
| 5 | UVR | - BpUVR8 | - B. platyphylla | - ABA treatment - Salt stress | - 10 μM ABA for 24 h - 100 mM NaCl for 24 h | - nucleus | - BpUVR8, as a specific receptor for UV-B, is regulated by salt stress and ABA treatment. It is also suggested that the UVR8 gene may be involved in the early signal transduction of salinity and ABA stress in addition to the process of UV-B signal transduction. | (Li et al. 2018) |
| MYB: Myeloblastosis; miRNAs: microRNAs; NAC: No apical meristem (NAM ), Arabidopsis transcription activation factor (ATAF1/2), Cup-shaped cotyledon (CUC2); UVR: UV resistance; BplMYB46; AHL: AT-Hook Motif Nuclear Localized; DHNs: Dehydrin-1; BpNACs: Betula platyphylla No apical meristem (NAM); BpUVR8: B. platyphylla UV resistance locus 8 gene; ABA: Abscisic acid; NaCl: Sodium chloride; MBS: MYB binding site involved in drought-inducibility; ABRE: Abscisic acid-responsive; N.A.: no detailed comments. | ||||||||
3.4 Drought stress
Many ERF and MYB family TFs were differentially regulated under drought stress (Wen et al. 2019). As BpERF2 and BpMYB102 are the examples of ERF and MYB TFs family that is involved in drought tolerance in B. platyphylla. BpERF2 overexpression significantly induced numerous genes involved in abiotic stress tolerance, illustrating that these genes are directly or indirectly regulated by BpERF2. Using ChIP-qPCR, it was revealed that BpERF2 could bind to the promoters of the birch genes homologous including LEA1, LEA8, LEA-D29, Dehydrin 2, heat shock protein-like (HSPL), HSPs, Root Primordium Defective 1 (RPD1), RD22-2, Pathogenesis-related Protein 1 (PRP1), and Beta-galactosidase (Wen et al. 2019).
The BpARF1 expression was induced by drought stress. The presence of the cis-acting elements in the BpARF gene promoters, resulting in drought stress resistance. The exon/intron structures analysis of BpARF genes revealed that the number of exons varied from 2 to 15 and all BpARF genes contained introns. Taken together, the BpARF members had similar exon/intron distribution within the same code group in gene structure, while those coded in other groups exhibited significant differences in gene structure. Moreover, it was found that binding sites responsive to drought frequently occurred in the B. platyphylla ARF family than those in phytohormone response, light responses, and meristem expression, via the PlantCARE database (Li et al. 2020a).
3.5 Heavy metal stress
Cd is one of the most critical global environmental pollutants, which inhibits plant growth and development and disturbs plants’ physiological processes. A previous study revealed that two lncRNAs namely LncRNA28068.1 and LncRNA30505.2 contributed to Cd tolerance in B. platyphylla. Heat shock protein 18.1 (HSP18.1) and L-lactate dehydrogenase A (LDHA) are the two lncRNAs target genes, which is enhanced Cd tolerance in B. platyphylla. Further experiment showed that the physiological and biochemical parameter such as EL, ROS content, and MDA value were significantly lower in HSP18.1 and LDHA OE lines under Cd stress, suggesting that lncRNAs participates in up-regulation or down-regulation of their target genes to improve Cd tolerance (Wen et al. 2020).
3.6 UV-B radiation
BpUVR8 is a specific receptor for UV-B. Nonetheless, hormonal signals and other abiotic stress such as drought, low temperature, limited N, and heat stress could regulate BpUVR8. BpUVR8 is involved in the early signal transduction of abiotic stresses and hormonal signals to process UV-B signal transduction. Multiple light response elements were found in the BpUVR8 promoter, which is appropriate with the BpUVR8 role in the light-induced photomorphogenesis pathway. Meanwhile, several tissue-specific cis-elements of hormone stress response including abscisic acid response element (ABRE), auxin response element (CATATG-motif), and jasmonic acid methyl ester response element (CGTCA-motif) were also found in the BpUVR8 promotor sequence. Besides, several abiotic stress-induced cis-elements, such as drought-induced response elements (MBS), low temperature-induced response elements, heat stress-induced response elements, and anaerobic response elements were also found in the BpUVR8 promoter sequences. To verify the contribution of the ABA signaling transduction pathway to BpUVR8 expression at the molecular level, Li et al. (2018) identify the relation of the BpHY5 gene and ABA-related genes (BpMYB2, BpABI3, BpABI4, BpABI5, and BpDREB2A). The expression levels of BpHY5 ABA-related genes except BpMYB2 and BpABI3 were elevated under ABA treatment. These results demonstrated that the characteristics of stress induction were found in the BpUVR8 promoter. Also, low temperature, droughts, and ABA were assumed to contribute to the expression of BpUVR8 (Li et al. 2018).
4 Conclusion and future perspective
Abiotic stress affects plant growth, development, and yield production of B. platyphylla. To provide the best insight and understanding of this topic, most studies combined physiological, biochemical, and molecular analyses. The increased EL, water loss, stomatal aperture, and chlorophyll degradation, as well as the reduction of photosynthesis rate, are the physiological measurement results in B. platyphylla under abiotic stress. Meanwhile, the enhancement of ROS accumulation (H2O2 and O2–), antioxidant enzymes (POD and SOD), MDA, and proline content are the most frequent result for biochemical measurements. According to the present studies, MYB and AP2/ERF TF (BplMYB46, BpMYB102, BpERF13, and BpERF2) are the dominant TF common in the reviewed article list. However, other genes from other TF families that contribute to abiotic stress tolerance of B. platyphylla such as BpHOX2, BpHMG6, BpHSP9, BpUVR8, BpBZR1, BplERD15, BpNACs needs to be explored. The mechanism of B. platyphylla abiotic stress tolerance was shown in Fig. 2. Abiotic stress-induced the expression of TF-related abiotic stress, and then these TF genes bind to cis-acting elements, resulting in the high or low expression of genes involved in abiotic stress tolerance. The expression of these abiotic stress tolerance genes influences the physiological and biochemical response of B. platyphylla to increase abiotic stress tolerance. This study also highlights the candidate genes that might have roles in B. platyphylla under abiotic stress. Further information on genomics and transcriptomics, as well as proteomics and metabolomics study, is beneficial to determine abiotic stress tolerance mechanism in B. platyphylla.
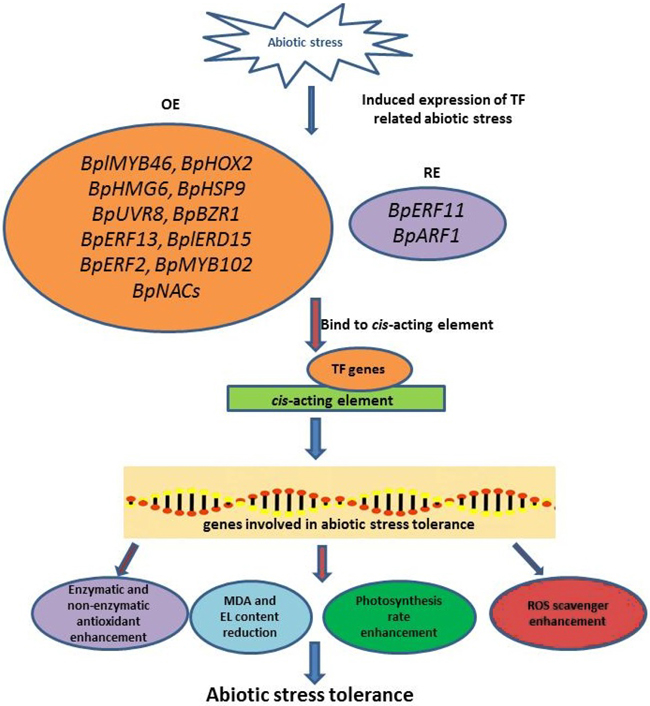
Fig. 2. The illustration of abiotic stress tolerance mechanism in Betula platyphylla. Abiotic stress-induced the expression of TF-related abiotic stress. Several TFs will be overexpressed or repress-expressed and followed binding to cis-acting elements to induce the expression of genes involved in abiotic stress tolerance. These genes involved in abiotic stress tolerance affects the plant response in physiological and biochemical aspect. BplMYB46: B. platyphylla myeloblastosis 46 gene; BpHOX2: B. platyphylla homeobox-leucine zipper 2 gene; BpHMG6: B. platyphylla high-mobility group 6 gene; BpHSP9: B. platyphylla heat shock protein 9 gene; BpUVR8: B. platyphylla UV resistance locus 8 gene; BpBZR1: B. platyphylla brassinazole-resistant 1 gene; BpERF13: B. platyphylla ethylene response factor 13 gene; BplERD15: B. platyphylla early response to dehydration 15 gene; BpERF2: B. platyphylla ethylene response factor 2 gene; BpMYB102: B. platyphylla myeloblastosis 102 gene; BpERF11: B. platyphylla ethylene responsive factor 11 gene; BpARF1: B. platyphylla auxin response factor 1 gene; BpNACs: B. platyphylla No apical meristem (NAM), Arabidopsis transcription activation factor (ATAF1/2), Cup-shaped cotyledon (CUC2) genes; MDA: Malondialdehyde; EL: Electrolyte leakage; ROS: Reactive oxygen species.
Acknowledgments
The authors appreciate the reviewers for comments and suggestions.
Author’s contributions
F.N.R. had contributed to writing and original draft preparation, J.N.N. edited the manuscript and interpreted the data and result, R.X.S interpreted the results, U.F, S.S, and A.T.L edited the manuscript, and S.C. contributed in supervision, project administration, funding acquisition, review, and editing manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This review was funded by the National Natural Science Foundation of China, grant number 31700579, and the Fundamental Research Funds for the Central Universities, grant number 2572019CG08.
Declaration of competing interest
The authors report no declarations of interest.
References
Akula R, Ravishankar GA (2011) Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal Behav 6: 1720–1731. https://doi.org/10.4161/psb.6.11.17613.
Ali RM, Abbas HM (2003) Response of salt stressed barley seedlings to phenylurea. Plant Soil Environ 49: 158–162. https://doi.org/10.17221/4107-PSE.
Alisoltani A, Karimi M, Ravash R, Fallahi H, Shiran B (2019) Molecular responses to cold stress in temperate fruit crops with focus on Rosaceae family. Genomics assisted breeding of crops for abiotic stress tolerance, vol II. Springer, Cham. https://doi.org/10.1007/978-3-319-99573-1_7.
Almeida T, Pinto G, Correia B, Santos C, Gonçalves S (2013) QsMYB1 expression is modulated in response to heat and drought stresses and during plant recovery in Quercus suber. Plant Physiol Bioch 73: 274–281. https://doi.org/10.1016/j.plaphy.2013.10.007.
An LJ, Ma Q, Du JX, Yu M, Li FR, Luan JY, Jiang J, Li HY (2019) Preliminary classification of the ABC transporter family in Betula halophila and expression patterns in response to exogenous phytohormones and abiotic stresses. Forests 10, article id 722. https://doi.org/10.3390/f10090722.
Arif Y, Singh P, Siddiqui H, Bajguz A, Hayat S (2020) Salinity induced physiological and biochemical changes in plants: an omic approach towards salt stress tolerance. Plant Physiol Bioch 156: 64–77. https://doi.org/10.1016/j.plaphy.2020.08.042.
Ashburner K, Mcallister HA (2013) The genus Betula, a taxonomic revision of birches. Royal Botanic Gardens, Kew.
Banik P, Zeng W, Tai H, Bizimungu B, Tanino K (2016) Effects of drought acclimation on drought stress resistance in potato (Solanum tuberosum L.) genotypes. Environ Exp Bot 126: 76–89. https://doi.org/10.1016/j.envexpbot.2016.01.008.
Bankaji I, Pérez-Clemente RM, Caçador I, Sleimi N (2019) Accumulation potential of Atriplex halimus to zinc and lead combined with NaCl: effects on physiological parameters and antioxidant enzymes activities. S Afr J Bot 123: 51–61. https://doi.org/10.1016/j.sajb.2019.02.011.
Beloiu M, Stahlmann R, Beierkuhnlein C (2020) High recovery of saplings after severe drought in temperate deciduous forests. Forests 11, article id 546. https://doi.org/10.3390/f11050546.
Chai L-Y, Wang Y, Yang Z-H, Mubarak H, Mirza N (2017) Physiological characteristics of Ficus tikoua under antimony stress. T Nonferr Metal Soc 27: 939–945. https://doi.org/10.1016/S1003-6326(17)60106-7.
Chen S, Lin X, Zhang D, Li Q, Zhao X, Chen S (2019) Genome-wide analysis of NAC gene family in Betula pendula. Forests 10, article id 741. https://doi.org/10.3390/f10090741.
Chen S, Wang Y, Yu L, Zheng T, Wang S, Yue Z, Jiang J, Kumari S, Zheng C, Tang H, Li J, Li Y, Chen J, Zhang W, Kuang H, Robertson JS, Zhao PX, Li H, Shu S, Yordanov YS, Huang H, Goodstein DM, Gai Y, Qi Q, Min J, Xu C, Wang S, Qu G-Z, Paterson AH, Sankoff D, Wei H, Liu G, Yang C (2021) Genome sequence and evolution of Betula platyphylla. Hortic Res 8, article id 37. https://doi.org/10.1038/s41438-021-00481-7.
Christie PJ, Alfenito MR, Walbot V (1994) Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 194: 541–549. https://doi.org/10.1007/BF00714468.
Chun HJ, Baek D, Cho HM, Lee SH, Jin BJ, Yun D-J, Hong Y-S, Kim MC (2019) Lignin biosynthesis genes play critical roles in the adaptation of Arabidopsis plants to high-salt stress. Plant Signal Behav 14, article id 1625697. https://doi.org/10.1080/15592324.2019.1625697.
Ciarkowska A, Ostrowski M, Jakubowska A (2016) Abiotic stress and phytohormones affect enzymic activity of 1-O-(indole-3-acetyl)-β-d-glucose: myo-inositol indoleacetyl transferase from rice (Oryza sativa). J Plant Physiol 205: 93–96. https://doi.org/10.1016/j.jplph.2016.07.018.
Cocozza C, Lasserre B, Giovannelli A, Castro G, Fragnelli G, Tognetti R (2009) Low temperature induces different cold sensitivity in two poplar clones (Populus×canadensis Mönch ‘I-214’ and P. deltoides Marsh.‘Dvina’). J Exp Bot 60: 3655–3664. https://doi.org/10.1093/jxb/erp212.
Deng X, Wang J, Li Y, Wu S, Yang S, Chao J, Chen Y, Zhang S, Shi M, Tian W (2018) Comparative transcriptome analysis reveals phytohormone signalings, heat shock module and ROS scavenger mediate the cold-tolerance of rubber tree. Sci Rep 8: 1–16. https://doi.org/10.1038/s41598-018-23094-y.
Dreyer A, Dietz K-J (2018) Reactive oxygen species and the redox-regulatory network in cold stress acclimation. Antioxidants 7, article id 169. https://doi.org/10.3390/antiox7110169.
Dubey AK, Khatri K, Jha B, Rathore MS (2021) The novel galactosyl transferase-like (SbGalT) gene from Salicornia brachiata maintains photosynthesis and enhances abiotic stress tolerance in transgenic tobacco. Gene 786, article id 145597. https://doi.org/10.1016/j.gene.2021.145597.
Eom HJ, Kang HR, Kim HK, Jung EB, Park HB, Kang KS, Kim KH (2016) Bioactivity-guided isolation of antioxidant triterpenoids from Betula platyphylla var. japonica bark. Bioorg Chem 66: 97–101. https://doi.org/10.1016/j.bioorg.2016.04.001.
Estravis-Barcala M, Mattera MG, Soliani C, Bellora N, Opgenoorth L, Heer K, Arana MV (2020) Molecular bases of responses to abiotic stress in trees. J Exp Bot 71: 3765–3779. https://doi.org/10.1093/jxb/erz532.
Fan G, Liu Y, Wang X, Zhan Y (2014) Cross-talk of polyamines and nitric oxide in endophytic fungus-induced betulin production in Betula platyphylla plantlets. Trees 28: 635–641. https://doi.org/10.1007/s00468-014-0978-1.
Feng J-P, Shi Q-H, Wang X-F (2009) Effects of exogenous silicon on photosynthetic capacity and antioxidant enzyme activities in chloroplast of cucumber seedlings under excess manganese. Agric Sci China 8: 40–50. https://doi.org/10.1016/S1671-2927(09)60007-9.
Gang H, Li R, Zhao Y, Liu G, Chen S, Jiang J (2019a) The birch GLK1 transcription factor mutant reveals new insights in chlorophyll biosynthesis and chloroplast development. J Exp Bot 70: 3125–3138. https://doi.org/10.1093/jxb/erz128.
Gang HX, Liu GF, Zhang MM, Zhao YM, Jiang J, Chen S (2019b) Comprehensive characterization of T-DNA integration induced chromosomal rearrangement in a birch T-DNA mutant. BMC Genomics 20, article id 311. https://doi.org/10.1186/s12864-019-5636-y.
Germanò MP, Cacciola F, Donato P, Dugo P, Certo G, D’angelo V, Mondello L, Rapisarda A (2012) Betula pendula leaves: polyphenolic characterization and potential innovative use in skin whitening products. Fitoterapia 83: 877–882. https://doi.org/10.1016/j.fitote.2012.03.021.
Guerriero G, Berni R, Muñoz-Sanchez JA, Apone F, Abdel-Salam EM, Qahtan AA, Alatar AA, Cantini C, Cai G, Hausman J-F, Siddiqui KS, Hernández-Sotomayor SMT, Faisal M (2018) Production of plant secondary metabolites: examples, tips and suggestions for biotechnologists. Genes 9, article id 309. https://doi.org/10.3390/genes9060309.
Guo H, Cui Z, Zhang Y, Wang C (2017a) Sequence characterization and expression analysis of NAC genes from Betula platyphylla. Trees 31: 1919–1931. https://doi.org/10.1007/s00468-017-1596-5.
Guo H, Wang Y, Wang L, Hu P, Wang Y, Jia Y, Zhang C, Zhang Y, Zhang Y, Wang C (2017b) Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol J 15: 107–121. https://doi.org/10.1111/pbi.12595.
Guo H, Wang L, Yang C, Zhang Y, Zhang C, Wang C (2018) Identification of novel cis‐elements bound by BplMYB46 involved in abiotic stress responses and secondary wall deposition. J Integr Plant Biol 60: 1000–1014. https://doi.org/10.1111/jipb.12671.
Hannus S, Hirons A, Baxter T, Mcallister HA, Wiström B, Sjöman H (2021) Intraspecific drought tolerance of Betula pendula genotypes: an evaluation using leaf turgor loss in a botanical collection. Trees 35: 569–581. https://doi.org/10.1007/s00468-020-02059-7.
He M, He CQ, Ding NZ (2018) Abiotic Stresses: General Defenses of Land Plants and Chances for Engineering Multistress Tolerance. Front Plant Sci 9.https://doi.org/10.3389/fpls.2018.01771.
Hoshika Y, Tatsuda S, Watanabe M, Wang X-N, Watanabe Y, Saito H, Koike T (2013a) Effect of ambient ozone at the somma of Lake Mashu on growth and leaf gas exchange in Betula ermanii and Betula platyphylla var. japonica. Environ Exp Bot 90: 12–16. https://doi.org/10.1016/j.envexpbot.2012.11.003.
Hoshika Y, Watanabe M, Inada N, Mao Q, Koike T (2013b) Photosynthetic response of early and late leaves of white birch (Betula platyphylla var. japonica) grown under free-air ozone exposure. Environ Pollut 182: 242–247. https://doi.org/10.1016/j.envpol.2013.07.033.
Hu P, Zhang K, Yang C (2019) BpNAC012 positively regulates abiotic stress responses and secondary wall biosynthesis. Plant Physiol 179: 700–717. https://doi.org/10.1104/pp.18.01167.
Huh JE, Hong JM, Baek YH, Lee JD, Choi DY, Park DS (2011) Anti-inflammatory and anti-nociceptive effect of Betula platyphylla var. japonica in human interleukin-1β-stimulated fibroblast-like synoviocytes and in experimental animal models. J Ethnopharmacol 135, 126–134. https://doi.org/10.1016/j.jep.2011.03.005.
Hussain SS (2019) Toward understanding the regulation of photosynthesis under abiotic stresses: recent developments. Photosynthesis, productivity and environmental stress. Wiley Online Library. https://doi.org/10.1002/9781119501800.ch8.
Iason GR, Taylor J, Helfer S (2018) Community-based biotic effects as determinants of tree resistance to pests and pathogens. For Ecol Manage 417: 301–312. https://doi.org/10.1016/j.foreco.2018.01.037.
IPNI (1911) International plant name index. Trav Mus Bot Acad Petersb. https://www.ipni.org/n/107857-1. Accessed 6 May 2021.
Ju EM, Lee SE, Hwang HJ, Kim JH (2004) Antioxidant and anticancer activity of extract from Betula platyphylla var. japonica. Life Sci 74: 1013–1026. https://doi.org/10.1016/j.lfs.2003.07.025.
Kamanga RM, Echigo K, Yodoya K, Mekawy AMM, Ueda A (2020) Salinity acclimation ameliorates salt stress in tomato (Solanum lycopersicum L.) seedlings by triggering a cascade of physiological processes in the leaves. Sci Hortic 270, article id 109434. https://doi.org/10.1016/j.scienta.2020.109434.
Kargiotidou A, Kappas I, Tsaftaris A, Galanopoulou D, Farmaki T (2010) Cold acclimation and low temperature resistance in cotton: Gossypium hirsutum phospholipase Dα isoforms are differentially regulated by temperature and light. J Exp Bot 61: 2991–3002. https://doi.org/10.1093/jxb/erq124.
Keinänen M, Julkunen-Tiitto R (1998) High-performance liquid chromatographic determination of flavonoids in Betula pendula and Betula pubescens leaves. J Chromatogr 793: 370–377. https://doi.org/10.1016/S0021-9673(97)00900-X.
Keinänen M, Julkunen-Tiitto R, Mutikainen P, Walls M, Ovaska J, Vapaavuori E (1999) Trade‐offs in phenolic metabolism of silver birch: effects of fertilization, defoliation, and genotype. Ecology 80: 1970–1986. https://doi.org/10.1890/0012-9658(1999)080[1970:TOIPMO]2.0.CO;2.
Khan MN, Siddiqui MH, Alsolami MA, Alamri S, Hu Y, Ali HM, Al-Amri AA, Alsubaie QD, Al-Munqedhi BMA, Al-Ghamdi A (2020) Crosstalk of hydrogen sulfide and nitric oxide requires calcium to mitigate impaired photosynthesis under cadmium stress by activating defense mechanisms in Vigna radiata. Plant Physiol Biochem 156: 278–290. https://doi.org/10.1016/j.plaphy.2020.09.017.
Kinose Y, Azuchi F, Uehara Y, Kanomata T, Kobayashi A, Yamaguchi M, Izuta T (2014) Modeling of stomatal conductance to estimate stomatal ozone uptake by Fagus crenata, Quercus serrata, Quercus mongolica var. crispula and Betula platyphylla. Environ Pollut 194: 235–245. https://doi.org/10.1016/j.envpol.2014.07.030.
Kitao M, Koike T, Tobita H, Maruyama Y (2005) Elevated CO2 and limited nitrogen nutrition can restrict excitation energy dissipation in photosystem II of Japanese white birch (Betula platyphylla var. japonica) leaves. Physiol Plant 125: 64–73. https://doi.org/10.1111/j.1399-3054.2005.00540.x.
Kitao M, Tobita H, Kitaoka S, Harayama H, Yazaki K, Komatsu M, Agathokleous E, Koike T (2019) Light energy partitioning under various environmental stresses combined with elevated CO2 in three deciduous broadleaf tree species in Japan. Climate 7, article id 79. https://doi.org/10.3390/cli7060079.
Lei X, Liu Z, Li X, Tan B, Wu J, Gao C (2021) Screening and functional identification of salt tolerance HMG genes in Betula platyphylla. Environ Exp Bot 181, article id 104235. https://doi.org/10.1016/j.envexpbot.2020.104235.
Lemmetyinen J, Järvinen P, Keinonen K, Lännenpää M, Keinänen M, Pasonen H-L (2008) Birches. Compendium of transgenic crop plants. Wiley Online Library. https://doi.org/10.1002/9781405181099.k0906.
Li H, Zhang X, Tong B, Wang Y, Yang C (2020a) Expression analysis of the BpARF genes in Betula platyphylla under drought stress. Plant Physiol Bioch 148: 273–281. https://doi.org/10.1016/j.plaphy.2020.01.028.
Li X, Ma M, Shao W, Wang H, Fan R, Chen X, Wang X, Zhan Y, Zeng F (2018) Molecular cloning and functional analysis of a UV-B photoreceptor gene, BpUVR8 (UV Resistance Locus 8), from birch and its role in ABA response. Plant Sci 274: 294–308. https://doi.org/10.1016/j.plantsci.2018.06.006.
Li X, Li R, Wang C, Yu Q, Chen S, Jiang J, Liu G (2020b) Inhibition of BpEIN3 causes plaques in leaves of Betula platyphylla × B. pendula. Trees 34: 483–495. https://doi.org/10.1007/s00468-019-01930-6.
Li Z, Pei X, Yin S, Lang X, Zhao X, Qu G-Z (2019) Plant hormone treatments to alleviate the effects of salt stress on germination of Betula platyphylla seeds. J For Res 30: 779–787. https://doi.org/10.1007/s11676-018-0661-2.
Liu Z, Wang P, Zhang T, Li Y, Wang Y, Gao C (2018) Comprehensive analysis of BpHSP genes and their expression under heat stresses in Betula platyphylla. Environ Exp Bot 152: 167–176. https://doi.org/10.1016/j.envexpbot.2018.04.011.
Lopez-Delacalle M, Silva CJ, Mestre TC, Martinez V, Blanco-Ulate B, Rivero RM (2020) Synchronization of proline, ascorbate and oxidative stress pathways under the combination of salinity and heat in tomato plants. Environ Exp Bot, article id 104351. https://doi.org/10.1016/j.envexpbot.2020.104351.
Lotfi N, Soleimani A, Vahdati K, Çakmakçı R (2019) Comprehensive biochemical insights into the seed germination of walnut under drought stress. Sci Hortic 250: 329–343. https://doi.org/https://doi.org/10.1016/j.scienta.2019.02.060.
Lv J, Li Y, Liu Z, Li X, Lei X, Gao C (2020a) Response of BpBZR genes to abiotic stress and hormone treatment in Betula platyphylla. Plant Physiol Biochem 151: 157–165. https://doi.org/10.1016/j.plaphy.2020.03.001.
Lv K, Li J, Zhao K, Chen S, Nie J, Zhang W, Liu G, Wei H (2019) Overexpression of an AP2/ERF family gene, BpERF13, in birch enhances cold tolerance through upregulating CBF genes and mitigating reactive oxygen species. Plant Sci 292, article id 110375. https://doi.org/10.1016/j.plantsci.2019.110375.
Lv K, Wei H, Jiang J (2020b) Overexpression of BplERD15 enhances drought tolerance in Betula platyphylla Suk. Forests 11, article id 978. https://doi.org/10.3390/f11090978.
Ma K, Jiang Y, Yu ZY, Huang YT, Zhan YG, Fan GZ (2019) H2S-induced NO/SNO positively promotes betulin production in Betula platyphylla. Ind Crops Prod 140, article id 111608.https://doi.org/10.1016/j.indcrop.2019.111608.
Mahajan M, Kuiry R, Pal PK (2020) Understanding the consequence of environmental stress for accumulation of secondary metabolites in medicinal and aromatic plants. J Appl Res Med Aroma 18, article id 100255. https://doi.org/10.1016/j.jarmap.2020.100255.
Malinowska M, Donnison I, Robson P (2020) Morphological and physiological traits that explain yield response to drought stress in miscanthus. Agronomy 10, article id 1194. https://doi.org/10.3390/agronomy10081194.
Mijiti M, Zhang Y, Zhang C, Wang Y (2017) Physiological and molecular responses of Betula platyphylla Suk to salt stress. Trees 31: 1653–1665. https://doi.org/10.1007/s00468-017-1576-9.
Mohri T, Mukai Y, Shinohara K (1997) Agrobacterium tumefaciens-mediated transformation of Japanese white birch (Betula platyphylla var. japonica). Plant Sci 127: 53–60. https://doi.org/10.1016/S0168-9452(97)00107-6.
Muraoka H, Koizumi H (2005) Photosynthetic and structural characteristics of canopy and shrub trees in a cool-temperate deciduous broadleaved forest: implication to the ecosystem carbon gain. Agric For Meteorol 134: 39–59. https://doi.org/10.1016/j.agrformet.2005.08.013.
Niinemets Ü (2010) Responses of forest trees to single and multiple environmental stresses from seedlings to mature plants: past stress history, stress interactions, tolerance and acclimation. For Ecol Manage 260: 1623–1639. https://doi.org/10.1016/j.foreco.2010.07.054.
Nowakowska JA, Stocki M, Stocka N, Ślusarski S, Tkaczyk M, Caetano JM, Tulik M, Hsiang T, Oszako T (2020) Interactions between Phytophthora cactorum, Armillaria gallica and Betula pendula Roth. seedlings subjected to defoliation. Forests 11, article id 1107. https://doi.org/10.3390/f11101107.
Oberschelp GPJ, Guarnaschelli AB, Teson N, Harrand L, Podestá FE, Margarit E (2020) Cold acclimation and freezing tolerance in three Eucalyptus species: a metabolomic and proteomic approach. Plant Physiol Biochem 154: 316–327. https://doi.org/10.1016/j.plaphy.2020.05.026.
Oh SR, Um JY, Choi HJ, Im CK, Kim KJ, Jung JW, Jeong GS, Hong SH, Kim SJ (2012) Betula platyphylla attenuated mast cell-mediated allergic inflammation in vivo and in vitro. Life Sci 91: 20–28. https://doi.org/10.1016/j.lfs.2012.05.018.
Pan Y, Niu M, Liang J, Lin E, Tong Z, Zhang J (2017a) Identification of heat-responsive miRNAs to reveal the miRNA-mediated regulatory network of heat stress response in Betula luminifera. Trees 31: 1635–1652. https://doi.org/10.1007/s00468-017-1575-x.
Pan Y, Niu MY, Liang JS, Lin EP, Tong ZK, Zhang JH (2017b) Identification of heat-responsive miRNAs to reveal the miRNA-mediated regulatory network of heat stress response in Betula luminifera. Trees 31: 1635–1652. https://doi.org/10.1007/s00468-017-1575-x.
Panche AN, Diwan AD, Chandra SR (2016) Flavonoids: an overview. J Nutr Sci 5: e47-e47. https://doi.org/10.1017/jns.2016.41.
Pellegrini E, Hoshika Y, Dusart N, Cotrozzi L, Gérard J, Nali C, Vaultier M-N, Jolivet Y, Lorenzini G, Paoletti E (2019) Antioxidative responses of three oak species under ozone and water stress conditions. Sci Total Environ 647: 390–399. https://doi.org/10.1016/j.scitotenv.2018.07.413.
Perea R, López-Sánchez A, Pallarés J, Gordaliza GG, González-Doncel I, Gil L, Rodríguez-Calcerrada J (2020) Tree recruitment in a drought- and herbivory-stressed oak-beech forest: implications for future species coexistence. For Ecol Manage 477, article id 118489. https://doi.org/10.1016/j.foreco.2020.118489.
Popov SA, Sheremet OP, Kornaukhova LM, Grazhdannikov AE, Shults EE (2017) An approach to effective green extraction of triterpenoids from outer birch bark using ethyl acetate with extractant recycle. Ind Crops Prod 102: 122–132. https://doi.org/10.1016/j.indcrop.2017.03.020.
Razieh JH, Vahide P, Najmeh AC, Kamal GB (2018) Improved accumulation of betulin and betulinic acid in cell suspension culture of Betula pendula roth by abiotic and biotic elicitors. Prep Biochem Biotechnol 48: 867–876. https://doi.org/10.1080/10826068.2018.1514514.
Riikonen J, Kivimäenpää M, Ossipov V, Saunier A, Marquardt P (2020) Metabolite composition of paper birch buds after eleven growing seasons of exposure to elevated CO2 and O3. Forests 11, article id 330. https://doi.org/10.3390/f11030330.
Ritonga FN, Chen S (2020) Physiological and molecular mechanism involved in cold stress tolerance in plants. Plants 9, article id 13. https://doi.org/10.3390/plants9050560.
Ritonga FN, Dwiyanti FG, Kusmana C, Siregar UJ, Siregar IZ (2018) Population genetics and ecology of Sumatran camphor (Dryobalanops aromatica) in natural and community-owned forests in Indonesia. Biodiversitas 19: 2175–2182. https://doi.org/10.13057/biodiv/d190625.
Shao FJ, Zhang L, Wilson IW, Qiu DY (2018) Transcriptomic analysis of Betula halophila in response to salt stress. Int J Mol Sci 19, article id 3412. https://doi.org/10.3390/ijms19113412.
Shaw K, Stritch L, Rivers M, Roy S, Wilson B, Govaerts R (2015) The red list of Betulaceae. Botanic Gardens Conservation International, Richmond, UK. ISBN 1-905154-58-0.
Šircelj H, Tausz M, Grill D, Batič F (2007) Detecting different levels of drought stress in apple trees (Malus domestica Borkh.) with selected biochemical and physiological parameters. Sci Hortic 113: 362–369. https://doi.org/10.1016/j.scienta.2007.04.012.
Su Z, Ma X, Guo HH, Sukiran NL, Guo B, Assmann SM, Ma H (2013) Flower development under drought stress: morphological and transcriptomic analyses reveal acute responses and long-term acclimation in Arabidopsis. Plant Cell 25: 3785–3807. https://doi.org/10.1105/tpc.113.115428.
Sun S, Kang XP, Tian YS, Zheng SW, Hao RJ, Liu QL, Zhang JC, Xing GM (2012) Molecular analysis of differentially expressed genes in birch (Betula Platyphylla) inflorescence. Biotechnol Biotec Eq 26: 2844–2854. https://doi.org/10.5504/BBEQ.2012.0012.
Tan Z, Wen X, Wang Y (2020) Betula platyphylla BpHOX2 transcription factor binds to different cis-acting elements and confers osmotic tolerance. J Integr Plant Biol 62: 1762–1779. https://doi.org/10.1111/jipb.12994.
Torun H, Novák O, Mikulík J, Pěnčík A, Strnad M, Ayaz FA (2020) Timing-dependent effects of salicylic acid treatment on phytohormonal changes, ROS regulation, and antioxidant defense in salinized barley (Hordeum vulgare L.). Sci Rep 10: 1–17. https://doi.org/10.1038/s41598-020-70807-3.
Wang QW, Qi L, Zhou WM, Liu C, Yu DP, Dai LM (2018) Carbon dynamics in the deciduous broadleaf tree Erman’s birch (Betula ermanii) at the subalpine treeline on Changbai Mountain, Northeast China. Am J Bot 105: 42–49. https://doi.org/10.1002/ajb2.1006.
Wang S, Huang HJ, Han R, Liu C, Qiu ZN, Liu GF, Chen S, Jiang J (2019a) Negative feedback loop between BpAP1 and BpPI/BpDEF heterodimer in Betula platyphylla × B. pendula. Plant Sci 289, article id 110280. https://doi.org/10.1016/j.plantsci.2019.110280.
Wang YM, Wang C, Guo HY, Wang YC (2019b) BplMYB46 from Betula platyphylla can form homodimers and heterodimers and is involved in salt and osmotic stresses. Int J Mol Sci 20, article id 1171. https://doi.org/10.3390/ijms20051171.
WCSP (2013) World checklist of selected plant families. http://apps.kew.org/wcsp/. Accessed 6 May 2021.
Wen X, Ding Y, Tan Z, Wang J, Zhang D, Wang Y (2020) Identification and characterization of cadmium stress-related LncRNAs from Betula platyphylla. Plant Sci 299, article id 110601. https://doi.org/10.1016/j.plantsci.2020.110601.
Wen XJ, Wang JX, Zhang DY, Wang YC (2019) A gene regulatory network controlled by BpERF2 and BpMYB102 in Birch under drought conditions. Int J Mol Sci 20, article id 3071. https://doi.org/10.3390/ijms20123071.
Yan S, Zhang DW, Chen S, Chen S (2020) Transcriptome Sequencing Analysis of Birch (Betula platyphylla Sukaczev) under Low-Temperature Stress. Forests 11, article id 970. https://doi.org/10.3390/f11090970.
Yin J, Li Y, Li C, Xiao J, Yang J, Li X, Sun L, Wang S, Tian H, Zhan Y (2020) Cloning, expression characteristics of a new FPS gene from birch (Betula platyphylla suk.) and functional identification in triterpenoid synthesis. Ind Crops Prod 154, article id 112591. https://doi.org/10.1016/j.indcrop.2020.112591.
Zhang MJ, Liu YL, Han GL, Zhang Y, Wang BS, Chen M (2020) Salt tolerance mechanisms in trees: research progress. Trees 35: 717–730. https://doi.org/10.1007/s00468-020-02060-0.
Zhang WH, Yang GY, Mu D, Li HY, Zang DD, Xu HY, Zou XZ, Wang YC (2016) An ethylene-responsive factor BpERF11 negatively modulates salt and osmotic tolerance in Betula platyphylla. Sci Rep 6, article id 23085. https://doi.org/10.1038/srep23085.
Zhao X, Guo P, Peng H, Zhao P, Yang Y, Zhang Z (2019) Potential of pulp production from whole-tree wood of Betula platyphylla Roth based on wood characteristics. Bioresources 14: 7015–7024.
Zhao XY, Bian XY, Li ZX, Wang XW, Yang CJ, Liu GF, Jiang J, Kentbayev Y, Kentbayeva B, Yang CP (2014) Genetic stability analysis of introduced Betula pendula, Betula kirghisorum, and Betula pubescens families in saline-alkali soil of northeastern China. Scand J For Res 29: 639–649. https://doi.org/10.1080/02827581.2014.960892.
Zhou LX, Yarra R, Jin LF, Cao HX (2020) Genome-wide identification and expression analysis of MYB gene family in oil palm (Elaeis guineensis Jacq.) under abiotic stress conditions. Environ Exp Bot 180, article id 104245. https://doi.org/10.1016/j.envexpbot.2020.104245.
Total of 101 references.

