Genetic structure of Tetraclinis articulata, an endangered conifer of the western Mediterranean basin
Sánchez-Gómez P., Jiménez J. F., Vera J. B., Sánchez-Saorín F. J., Martínez J. F., Buhagiar J. (2013). Genetic structure of Tetraclinis articulata, an endangered conifer of the western Mediterranean basin. Silva Fennica vol. 47 no. 5 article id 1073. https://doi.org/10.14214/sf.1073
Highlights
- The employment of ISSR molecular markers has shown moderate genetic diversity and high genetic differentiation in Tetraclinis articulata
- Genetic structure of populations seems to be influenced by the anthropogenic use of this species since historical times, or alternatively, by the complex palaeogeographic history of the Mediterranean basin
- Results could be used to propose management policies for conservation of populations.
Abstract
Tetraclinis articulata (Vahl) Masters is a tree distributed throughout the western Mediterranean basin. It is included in the IUCN (International Union for Conservation of Nature) red list, and protected by law in several of the countries where it grows. In this study we examined the genetic diversity and genetic structure of 14 populations of T. articulata in its whole geographic range using ISSR (inter simple sequence repeat) markers. T. articulata showed moderate genetic diversity at intrapopulation level and high genetic differentiation. The distribution of genetic diversity among populations did not exhibit a linear pattern related to geographic distances, since all analyses (principal coordinate analysis, Unweighted pair group method with arithmetic mean dendrogram and Bayesian structure analysis) revealed that spanish population grouped with Malta and Tunisia populations. Although it is possible that T. articulata earlier was natural in Southeast Spain, results suggest that the current population has been reintroduced into the Iberian Peninsula in historical times, due to its utility in mining and building. In addition, results could be used to propose management guidelines for the conservation of T. articulata.
Keywords
conservation;
genetic differentiation;
genetic diversity;
ISSR markers
- Sánchez-Gómez, Departamento de Biología Vegetal (Botánica), Universidad de Murcia, Campus de Espinardo s/n, E-30100 Murcia, Spain E-mail psgomez@um.es
-
Jiménez,
Departamento de Biología Vegetal (Botánica), Universidad de Murcia, Campus de Espinardo s/n, E-30100 Murcia, Spain
E-mail
fjimenez@um.es

- Vera, Departamento de Biología Vegetal (Botánica), Universidad de Murcia, Campus de Espinardo s/n, E-30100 Murcia, Spain E-mail jbveraperez@gmail.com
- Sánchez-Saorín, Dirección General de Medio Ambiente, Consejería de Presidencia de la Región de Murcia, C/ Catedrático Eugenio Úbeda nº3, E-30071 Murcia, Spain E-mail fjavier.sanchez3@carm.es
- Martínez, Dirección General de Medio Ambiente, Consejería de Presidencia de la Región de Murcia, C/ Catedrático Eugenio Úbeda nº3, E-30071 Murcia, Spain E-mail juanf.martinez@carm.es
- Buhagiar, Argotti Herbarium and Gardens (UOM), Triq Vincenzo Bugeja Floriana VLT 16, Malta E-mail joseph.buhagiar@um.edu.mt
Received 18 March 2013 Accepted 2 December 2013 Published 16 December 2013
Views 194910
Available at https://doi.org/10.14214/sf.1073 | Download PDF

1 Introduction
The genus Tetraclinis Masters comprises only one species, T. articulata (Vahl) Masters, which belongs to the family Cupressaceae. Tetraclinis articulata was initially included in the genus Thuja L., which is currently well represented in Asia and North America. It has also been included in the genus Callitris Vent., which is now considered endemic to Oceania, and has also been related to the South African genus Widdringtonia Endl (Farjon 2010).
At present it is to be found in a restricted area, covering approximately one million hectares. There are obvious discontinuities in population distribution, indicating the ancient and relict nature of this taxon. Its range mainly includes Morocco, Algeria and Tunisia (Quézel 1980). Besides these main populations, the species can be found in low numbers of individuals on the island of Malta and in south-eastern Spain. The latter location (Sierra de Cartagena) is thought to be the only natural enclave of this species in continental Europe.
According to Quézel (1980) T. articulata is a species whose optimum altitudes range from sea level up to 1000–1100 m. It grows on shady slopes and even at altitudes of 1500–1800 m on south-facing slopes, depending on the latitude. Tetraclinis. articulata lives mainly in carbonate soils; however, it can also be found in soils ranging from calcareous and dolomites to granites, rhyolites and shales. Noteworthy is its presence in toxic soils, rich in zinc and lead (calamine). This species can live in semiarid ombroclimatic zones (250 mm annual precipitation) as well as in clearly humid ombroclimatic areas, like some parts of Morocco and the Algerian coast (Benabid 1984; Quézel 1980). Moreover, Fennane et al. (1984) indicate that T. articulata may be favoured by fog and sea mist, which usually represent an ecological optimum for other more demanding species.
Although this species is interesting from the scientific and conservation viewpoint, its intraspecific genetic variation remains unknown. Studies into the genetic structure of plant species are increasingly common in conservation and recent decades have seen several significant studies demonstrating the distribution of genetic variation in rare and endangered species (Falk and Holsinger 1991; Ellstrand and Elam 1993). Genetic diversity is directly involved in species viability. Therefore, loss of genetic diversity may lead to a decrease in population viability, adaptability to environmental changes, and disease resistance (Beardmore 1983; Fischer and Matthies 1998; Reed and Frankham 2003; Frankham et al. 2010).
The genetic structure of populations is usually related to intrinsic factors, such as mating system and gene flow among populations, and extrinsic factors, such as population ecology and significant historical events (bottlenecks, expansion or fragmentation). In conifers, a number of studies have revealed high genetic variation at intra-population level and low genetic differentiation among populations (Hamrick et al. 1992; Nybom and Bartish 2000). However, this distribution of genetic variation is not a general pattern, and there are numerous examples in the literature, for example: species that show low genetic diversity and high levels of genetic differentiation (Xiao et al. 2004; Ge et al. 2005) or high genetic diversity and moderate to high genetic differentiation (Jiménez et al. 2003; Boratynski et al. 2009).
Different molecular methods can be used to efficiently study the gene pool of rare species. In last 20 years, a technique based on DNA fingerprinting, the Intersimple Sequence Repeat (ISSR) method (Zietkiewicz et al. 1994) has frequently been used. ISSR involves the use of a single primer composed of short repeated sequences, di- tri- or tetra-nucleotides, plus a short arbitrary sequence (anchor), which targets a subset of SSRs (simple sequence repeats) or microsatellites and amplifies the region between two closely spaced and oppositely oriented simple sequence repeats. These microsatellites are abundant throughout the eukaryotic genome (Tautz and Renz 1984; Kijas et al. 1995) and evolve quickly (Levinson and Gutman 1987). This technique is relatively fast, cost-efficient and does not require prior knowledge of the genome.
The main objectives of this study were: i) to assess the level and distribution of T. articulata genetic variation; ii) to investigate the relationships among known populations by statistical and cluster analyses; and iii) to provide information useful to establish management and conservation guidelines of T. articulata populations.
2 Material and methods
2.1 Plant material
The sampling strategy covered the whole Tetraclinis articulata distribution range, so that material was collected from 14 populations belonging to Spain (2 populations), Morocco (5 populations), Algeria (3 populations), Tunisia (1 population) and Malta (3 populations). A total of 215 individuals were included in the study. The number of individuals sampled per population varied from 10 to 31 and the location of populations are shown in Table 1 and Fig. 1. Transects were made, collecting specimens sufficiently far apart (where was possible) to avoid collecting very closely related individuals. Only small branches (under 10 cm) were sampled to avoid damaging specimens. To avoid degradation of plant tissues, all plant material was labelled and kept in sealed bags with silica gel according to Sytsma et al. (1993) until DNA extraction.
| Table 1. Geographical location of fourteen sampled populations of Tetraclinis articulata and genetic diversity estimates. P percentage of polymorphic loci, H0 Nei’s heterozygosity (genetic diversity), H0* Nei’s heterozygosity calculated for a standardised population size of 10, Hs mean heterozygosity within populations, Ht total heterozygosity between populations, Gst index of population differentiation. | ||||||||
| Population | Location | Individuals sampled | (P) % polymorph. loci | H0 | H0* | Hs | Ht | Gst |
| Algeria | ||||||||
| 1 Zemmora | 38°42.80´N, 0°45.36´E | 10 | 48.06 | 0.159 | 0.159 | |||
| 2 El Fedjoudj | 36°30.40´N, 7°21.70´E | 10 | 45.37 | 0.178 | 0.178 | |||
| 3 Saida | 34°50.45´N, 0°04.63´E | 10 | 39.14 | 0.160 | 0.160 | |||
| Morocco | ||||||||
| 4 Khenifra | 33°05.69´N, 5°35.03´W | 10 | 37.40 | 0.153 | 0.153 | |||
| 5 El Ksiba | 32°34.35´N, 6°02.81´W | 21 | 64.63 | 0.206 | 0.201 | |||
| 6 Oujda | 34°30.92´N, 1°54.21´W | 20 | 60.98 | 0.184 | 0.174 | |||
| 7 Hassi Berkane | 34°43.60´N, 2°47.60´W | 10 | 45.07 | 0.154 | 0.154 | |||
| 8 Ain el Aouda | 33°44.66´N, 6°45.05´W | 21 | 68.29 | 0.204 | 0.195 | |||
| Malta | ||||||||
| 9 Wied Mizieb | 35°51.89´N, 14°24.17´E | 31 | 73.17 | 0.183 | 0.154 | |||
| 10 Argotti | 35°53.48´N, 14°30.11´E | 14 | 53.66 | 0.161 | 0.169 | |||
| 11 Gnien Ingraw | 35°57.71´N, 14°21.48´E | 20 | 79.27 | 0.231 | 0.205 | |||
| Spain | ||||||||
| 12 Cenizas | 37°35.49´N, 0°49.67´W | 14 | 69.51 | 0.227 | 0.195 | |||
| 13 Sabinar | 37°37.22´N, 0°47.87´W | 14 | 60.98 | 0.201 | 0.190 | |||
| Tunisia | ||||||||
| 14 Cape Bon | 37°04.27´N, 11°04.27´E | 10 | 47.56 | 0.151 | 0.151 | |||
| Total | 215 | 97.56 | 0.179 | 0.258 | 0.31 | |||
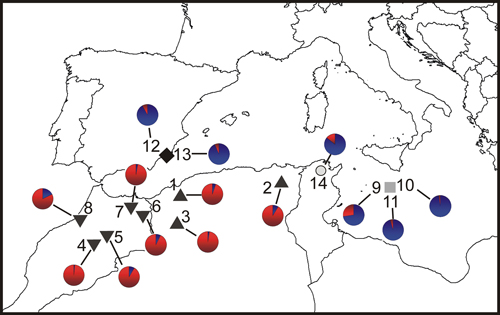
Fig. 1. Location map and Bayesian analysis of the genetic structure of 215 individuals from fourteen sampled populations of T. articulata. Pie charts represent the mean proportion of membership to the predefined K = 2 clusters with the highest ΔK obtained following Evanno et al. (2005). Inverted triangles, Morocco populations; triangles, Algeria populations; grey square, Malta populations; dark diamond, Spain populations; open circle, Tunisia population.
2.2 DNA extraction and amplification
The CTAB method (Doyle and Doyle 1987) was used to extract genomic DNA. The ISSR reactions were performed in 25 µl, containing 10 mM Tris-HCl (pH 8.8, 25 °C), 50 mM KCl, 1.5 mM MgCl2, 200 µM of each dATP, dCTP, dGTP and dTTP, 15 ng primer, approximately 25 ng genomic DNA and one unit of Taq polymerase. A control, containing all the components except genomic DNA, was included in each set of reactions to rule out possible contamination. Amplifications were carried out in an Eppendorf Thermocycler under the following conditions: an initial cycle at 94 °C for 2 min; 39 cycles of 30 sec at 94 °C, 30 sec at 47 °C, 1 min at 72°C. A final cycle at 72 °C for 10 min was included. Five ISSR primers were used (Table 2). These were chosen after testing 20 different ISSR primers. Duplicate amplifications were conducted for each sample in order to ensure reproducible results and minimize errors. Bands which could not be reproduced in both assays were not considered for analysis. Amplification products were run on 1.5% agarose gels in TAE buffer, stained with ethidium bromide and viewed under ultraviolet light.
| Table 2. ISSR primers used to generate multilocus profiles with DNA from T. articulata. | ||||
| Primer | Sequence | Range | No. bands | % polymorphic |
| OLIGO1 | (GT)6RG | 200–1220 | 20 | 100 |
| OLIGO4 | (GT)6AY | 190–1310 | 12 | 100 |
| UBC857 | (AC)8YG | 220–1405 | 18 | 100 |
| UBC881 | (GGGT)3G | 200–1550 | 16 | 87.5 |
| OW12 | (GA)8YA | 230–1330 | 16 | 100 |
2.3 Statistical analysis
Gel images were captured in a Kodak Gel Logic System, and fragment sizes were determined by comparison with molecular weight standards (GeneRuler 50 bp DNA ladder, Fermentas) using the Kodak 1D 3.6 software. The presence or absence of each ISSR fragment was treated as a binary character (coded 1 and 0, respectively) and used to construct the original data matrix. Following suggestions by Grosberg et al. (1996), no attempts were made to code for band intensity. Bands showing the same gel mobilities were assumed homologous. DNA bands showing quantitative variation in brightness were scored as present, regardless of their intensities, and absent if they were undetectable. A similarity matrix was generated with the 1/0 matrix, using Dice coefficient (Dice 1945) with the SPSS 13.0 program. The Dice coefficient takes into account the presence of shared and unique bands present in samples, but not the shared absence of bands (Sneath and Sokal 1973). This similarity matrix was used to perform a cluster analysis following the UPGMA-method (Unweighted Pair Group Method with Arithmetic Averages), and a Principal Coordinate Analysis (PCoA). These analyses were performed with Multivariate Statistical Package software (MVSP version 3.12d; http://www.kovcomp.com/mvsp).
Intra-specific genetic diversity was inferred with POPGENE 1.32 program (Yeh et al. 1999). The parameters estimated were a percentage of polymorphic loci, H0, (the diversity within populations), Hs (average gene diversity within populations) and Ht (Total gene diversity; Nei 1973). Additionally, to eliminate the influence of uneven sample sizes per population, H0 was calculated for a standardised population size of 10 randomly selected individuals. Genetic distance among populations (Gst) was computed following Nei (1973). Pairwise genetic distances among populations were also carried out (Nei 1972), and used to perform a UPGMA dendrogram.
Analysis of Molecular Variance (AMOVA, Excoffier et al. 1992) was conducted to estimate variance components at several hierarchical levels, partitioning the variation among predefined geographic groups (Spain, North Africa, Malta), among populations and among individuals within populations. Taking into account that ISSR markers are dominant, in order to estimate population genetic structure we made assumptions that null bands are homologous and that populations are in Hardy-Weinberg equilibrium (Lynch and Milligan 1994). In order to test the correlation between genetic and geographic distances (in kilometers) among populations, Mantel test was performed. The AMOVA analysis and Mantel test were undertaken with the Arlequin 2000 program (Schneider et al 2000).
A Bayesian model-based analysis was performed to infer population structure with Structure version 2.2 (Pritchard et al. 2000; Falush et al. 2007). The F model, based on an admixture ancestry model with correlated allele frequencies, was imposed to estimate the posterior probabilities [LnP(D)] of K groups (Pritchard and Wen 2004) and the individual percentages of membership assigned to them according to their molecular multilocus profiles (Falush et al. 2003; 2007). Probabilities for a range of K were examined starting from 1 to the number of sampled populations plus one (K = 1–15), using a burn-in period and run length of the Markov chain Monte Carlo (MCMC) of 105 and 106 iterations, respectively. Twenty runs were carried out for each K, and the rate of change in the log probability of data between successive likelihood values (ΔK) was estimated as the average of the 20 replicates following Evanno et al. (2005). According to these authors, the modal value of the distribution of each ΔK corresponds to the real K.
3 Results
3.1 Genetic diversity
The five selected primers generated 82 reproducible ISSR bands, ranging in size from 190 to 1550 bp (Table 2). Most of these bands were polymorphic (97.56%), and none of the 215 individuals shared the same phenotype. Unfortunately, no population markers were detected. The percentages of polymorphic loci (P) for a single population ranged from 79.27% (Gnien Ingraw, Malta) to 37.40% (Khenifra, Morocco). Within-population genetic diversity was in accordance with P values. Malta harboured the most diverse population (H0 = 0.231), whereas Cape Bon harboured the lowest within-population diversity (H0 = 0.151). In general, populations were not genetically impoverished, being the two Spanish populations, Ain el Aouda and El Ksiba (both from Morocco) and Gnien Ingraw (Malta) the most diverse. The standardised H0 values suggest that intrapopulation genetic diversity is not influenced by the number of individuals sampled. The average gene diversity within populations (Hs) was 0.179. The total diversity (Ht = 0.258) shows a high genetic diversity in the species, and the differentiation index (Gst = 0.31) shows a high genetic differentiation among populations (Table 1). Genetic distances between populations of T. articulata ranged from 0.031 (between El Ksiba and Oujda) to 0.226 (between Khenifra and Argotti)
AMOVA analysis showed that the greatest percentage of variation was attributable to within-population diversity (82.69%). Among populations within regions accounted for 7.88% of the variation, whereas only 9.44% of variation was attributable to predefined geographic groups. Mantel test revealed a significant but low correlation between genetic and geographic distances (r = 0.331, P = 0.002, 5000 permutations).
3.2 Population structure
The cluster phenogram obtained with the UPGMA algorithm did not group the individual specimens within their own populations nor with their own regional provenances (results not shown). The UPGMA dendrogram, inferred with the genetic distances among populations, showed two subclades (Fig. 2). The first one clustered the Moroccan and Algerian populations while the other clade clustered the populations from Malta, Tunisia and Spain. The PCoA showed similar results (Fig. 3). The variation explained by the first three axes accounted for 62.81% (38.44%, 18.04% and 6.33%, respectively). Although individual specimens did not form discrete groups of populations, the PCoA graph showed that specimens from Spain, Malta and Tunisia clustered together, whereas Moroccan and Algerian ones tended to form the other group.
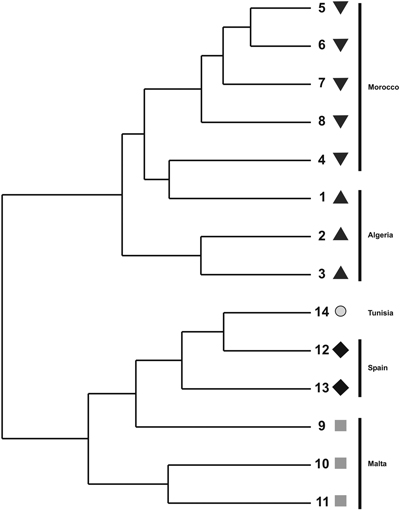
Fig. 2. UPGMA dendrogram constructed using pair-wise Nei’s (1973) genetic distance derived by ISSR analysis for populations of T. articulata.
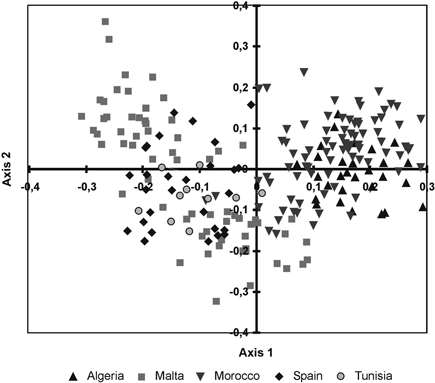
Fig. 3. Principal coordinate analysis of the 215 individuals from fourteen sampled populations of T. articulata based on pair-wise Nei’s (1973) genetic distances.
The Bayesian analysis of population genetic structure in Tetraclinis articulata conducted with Structure found the highest LnP(D) and ΔK values for K = 2. This grouping showed similar results to those obtained with the PCoA. Populations from Algeria and Morocco clustered together, differentiating them from populations growing in Spain, Malta and Tunisia (Fig. 1).
4 Discussion
4.1 Genetic diversity
Numerous studies have shown that conifers, as long-lived and outcrossing species, typically exhibit high levels of within-population genetic variation and low genetic differentiation (Hamrick et al. 1992; Nybom and Bartish 2000; Dzialuk et al. 2011). However, it is often difficult to predict variation patterns within the context of the complex historical, geological and climatic changes occurring in the Mediterranean. On considering values for Tetraclinis articulata genetic variation, one finds they are similar to those found in other conifers inhabiting the Mediterranean region (Fady-Welterlen 2005). Comparisons with other studies are difficult since genetic diversity depends on numerous factors, such as life history, breeding system, growth life forms, geographical range and even the type of molecular method used (Powell et al. 1996; Nybom 2004). In spite of these complications, if we compare the results of studies for other mediterranean conifers (Terrab et al. 2006; 2007; 2008; Douaihy et al. 2011; Dzialuk et al. 2011) T. articulata is not genetically impoverished. These genetic diversity values should be considered with caution. Although T. articulata is locally abundant in North Africa, it consists of fragmented populations due to anthropogenic impact (Quézel and Médail 2003), but this circumstance is not reflected in the within-population genetic diversity observed. This might be explained by two factors: 1) A recent fragmentation of populations or 2) the persistence of trees with high levels of heterozigosity in most of populations.
Regarding genetic differentiation, the fixation index (Gst) between pairs of T. articulata populations would indicate that, with the exception of some pairs of geographically close populations, there is often moderate to high genetic differentiation. Plant population genetic differentiation reflects interactions amongst a range of different processes, including the long-term evolutionary history of the species (e.g. shifts in distribution, habitat fragmentation and population isolation), mutation, genetic drift, mating systems, gene flow and selection (Schaal et al. 1998). As discussed earlier, T. articulata is located in fragmented and genetically differentiated populations, and in some cases affected by human use, which could be one of the reasons for the genetic differentiation observed, together with the geological reorganisation of the Western Mediterranean basin during the Oligocene-Pliocene ages (Magri et al. 2007, and references therein), and the impact of the climatic changes ocurred during Quaternary glaciations which resulted in a number of retraction and population recolonization processes of numerous species (Fady-Welterlen 2005).
4.2 Population structure
Under conditions in which there is a balance between genetic drift and gene flow, inter-population variation is often related to the geographical distance between populations (Pérez-Collazos et al. 2010); however, the Mantel test suggests that, at least partially, this pattern was not followed by Tetraclinis articulata. The North African populations (Morocco and Algeria) clustered together, as indicated by the PCoA, the UPGMA dendrogram, and the Bayesian structure analysis, while populations of Tunisia and Malta also clustered together. These locations might be in contact by a land bridge that connected Tunisia, Malta and Sicily during the Messinian salinity crisis. Up to this point, isolation by distance correlation pattern was apparent. However, the Spanish populations did not match this pattern. We could expect these populations to be more genetically similar to North African populations, given the proximity and the pattern followed by many species inhabiting Iberia-North Africa (Jiménez et al. 2003; Rodríguez-Sánchez et al. 2008). However, all analyses grouped the Spanish population with those from Malta and Tunisia. In fact, sometimes, the genetic structure of plant populations does not only correspond to geographical patterns but also reflects the interaction of different processes. Such processes include long-term and short-term evolutionary events, such as migration, diversification, habitat fragmentation, gene flow and selection (Slatkin and Barton 1989).
There has been much debate about the origins of Iberian T. articulata populations which are located exclusively in the coastal mountains of Murcia, locally known as “Sierra Minera” of Cartagena. Mining in these mountains dates back to Phoenician times and continues today, and T. articulata wood, which is very hard, was used to construct the mine shafts. Indeed, the use of T. articulata by man dates back to prehistoric times, with remains of charcoal and pollen having been found in numerous Southeast Iberian archaeological sites dating from the Palaeolithic to 1000 years ago (Carrión et al. 1995; García and Grau 2005; García et al. 2008). Its main anthropogenic use is related to construction, carpentry and fuel. Moreover, the bark is of value for tanning and the resin, known as Sandarac, has numerous industrial and medical applications.
Two alternative hypotheses could explain the genetic structure of T. articulata: 1) Iberian population appears to have been in contact with the easternmost populations, either by long-distance germplasm transport or by transfer of plant material from Tunisian stocks, probably for the purposes mentioned above, as suggested by data obtained with PCoA and Bayesian methods. However, high levels of genetic variation in this population (H = 0.236) do not indicate a founder effect mediated by the creation of a new population, which is the pattern observed with cpSSR and isozymes markers in ancient Cedrus libani A. Rich. reintroductions (Fady et al. 2008). The relationship between the populations in Southeast Spain, Malta and Tunisia, could be related to the use and spreading of the species since past times, especially given the trading activities of the Phoenicians, who were highly active in these regions. The fact that fossil records show prior presence in Spain does not contradict this hypothesis, as it is possible that indigenous populations waned in certain historical periods, but were later introduced by man in these regions of strategic economic importance. Indeed, it is noteworthy that there are only relict populations near the mining area of Cartagena, since the ecological optimum for this species is located in northern Africa.
2) The genetic affinity between Iberian, Tunisian and Maltese populations could reflect the complex palaeogeographic history of the Mediterranean basin. This hypothesis suggests a scenario in which after an initial expansion of T. articulata throughout the Mediterranean basin, western European populations (Iberian) contacted (or originated) the easternmost populations according to the tectonics of the western Mediterranean since the Oligocene. This could be possible because during the Oligocene, the European-Iberian continental assembled continental terranes that are now found in Balearic islands, Corsica, Sardinia, Sicily, Calabria, north of Algeria and Rif in Morocco, and during the Miocene drifted towards their current positions (Rosenbaum et al. 2002; Magri et al. 2007). These patterns have been found in Quercus suber L. (Magri et al. 2007), or Pinus pinaster Aiton (Burban and Petit 2003). However, accepting this hypothesis requires a) that Strait of Gibraltar would have acted as a barrier for gene flow (Terrab et al. 2008; Boratynski et al. 2013), remaining isolated Spanish and Moroccan populations, b) assuming the existence in historical times, of intermediate populations related to Spanish populations, maybe in the Mediterranean islands (e.g. Sicily, Sardinia...), or in northern Algeria, and c) long term persistence of current populations and stability of genetic variation (perhaps since Tertiary). Current populations of T. articulata are located near from several known glacial refugia (Médail and Diadema 2009). Moreover, T. articulata, as other Mediterranean palaeo-endemic species, shows low evolution rates in DNA (Comes 2004; Petit and Hampe 2006). In fact, in an attempt to establish phylogeographic relationships among populations using other molecular markers, we sequenced several chloroplast regions (trnT-trnL, trnL-F, trnT-trnD, trnE-trnY), that did not yield results, as identical sequences appeared in all populations (results not shown).
Conversely, although Juniperus thurifera L. showed a pattern of genetic structure that could match with this hypothesis, Terrab et al. (2008) suggested that genetic similarities between Algerian and Europe populations were due to several migration events from Europe, most probably via long-distance dispersals. The use of other more specific markers (e.g., microsatellite), as well as more intensive sampling of northern African populations, especially in Algeria, could shed some light and help validate these hypotheses.
4.3 Implications for conservation
As discussed, Tetraclinis articulata has been put to extensive use as a raw material. In general, T. articulata wood has been used to line mineshafts and, locally, in housing construction. It is still employed for most of these traditional uses in North African countries; however, it is currently a protected species in Spain and Malta, and its use is strictly regulated. The main threats facing T. articulata are related to overexploitation, overgrazing and habitat deterioration in northern Africa. Meanwhile, in Spain and Malta, habitats are very small and fragmented, close to human population and activity, and prone to invasion by exotic/invasive species, destruction by fire, and so on. Notwithstanding, T. articulata is listed as a “Least Concern” species within its habitat range (Sánchez-Gómez et al. 2011).
The ultimate goals of conservation are to ensure sustainable survival of populations and to preserve their evolutionary potential. The estimates of genetic diversity and genetic differentiation provide a basis for implementing efficient and practical conservation programs for Tetraclinis. articulata. This species has suffered drastic fragmentation and reduction of its populations throughout history. Forest fragmentation is considered a major factor, inducing the loss of genetic diversity (Ellstrand and Elam 1993; Young et al. 1996). Nevertheless, in the present study, genetic variability distribution analysis, as estimated by ISSRs, suggests that 83% of the total genetic variation is still harboured within populations. This leads to the next conclusion: the species is not endangered yet, and, for conservation purposes, it would probably be sufficient to maintain the populations located across the whole distribution range, to ensure continued representation of the total genetic diversity. Based on the levels of genetic differentiation between population pairs, population support strategies could be established in cases where population viability may be at risk due to a drastic reduction in the number of specimens.
Acknowledgements
This work was supported by grants from the Spanish Ministry of Education and Science (CGL2011-30099) and an agreement between the University of Murcia and the General Directorate of Environment of Autonomous Community of Murcia. Authors wish to thank to Jaime Güemes from University of Valencia, who collected samples of Tunisia, and Miloud Aouissat from University of Saida, who collected samples of Algeria.
References
Benabid A. (1984). Etude phytoecologique des peuplements forestiers et preforestiers du rif centro-occidental (Maroc). Travaux de l’Institut Scientifique, série Botanique, nº 34: 1–64.
Beardmore J.A. (1983). Extinction, survival, and genetic variation. In: Schoenwald-Cox C.M., Chambers S.M., MacBryde B., Thomas L. (eds.). Genetics and conservation. Benjamin-Cummings, California, Menlo Park. p. 125–151.
Boratynski A., Lewandowski A., Boratynska K., Montserrat J.M., Romo A. (2009). High level of genetic differentiation of Juniperus phoenicea (Cupressaceae) in the mediterranean region: geographic implications. Plant Systematics and Evolution 277: 163–172. http://dx.doi.org/10.1007/s00606-008-0122-z.
Boratynski A., Jasinska A.K., Marcysiak K., Mazur M., Romo A.M., Boratynska K., Sobierajska K., Iszkulo G. (2013). Morphological differentiation supports the genetic pattern of the geographic structure of Juniperus thurivera. Plant Systematics and Evolution 299: 773–784. http://dx.doi.org/10.1007/s00606-013-0760-7.
Burban C., Petit R.J. (2003). Phylogeography of maritime pine inferred with organelle markers having contrasted inheritance. Ecology 12: 1487–1495.
Carrión J.S., Munuera M., Dupré M. (1995). Estudios de Palinología arqueológica en el Sureste Ibérico semiárido. Cuaternario y Geomorfología 9(3–4): 17–31.
Comes H.P. (2004). The Mediterranean region – a hotspot for plant biogeographic research. New Phytologist 164: 11–14. http://dx.doi.org/10.1111/j.1469-8137.2004.01194.x.
Dice L.R. (1945). Measures of the amount of ecologic association between species. Ecology 26: 297–302. http://dx.doi.org/10.2307/1932409.
Douaihy B., Vendramin G.G., Boratynski A., Machon N., Dagher-Kharrat M.B. (2011). High genetic diversity with moderate differentiation in Juniperus excelsa from Lebanon and the eastern Mediterranean region. AoB Plants 01/2011. http://dx.doi.org/10.1093/aobpla/plr003.
Doyle J.J., Doyle J.L. (1987). A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochemical Bulletin 19: 11–15.
Dzialuk A., Mazur M., Boratynska K., Monserrat J.M., Romo A., Boratynski A. (2011). Population genetic structure of Juniperus phoenicea (Cupressaceae) in the western Mediterranean Basin: gradient of diversity on a broad geographical scale. Annals of Forest Science 68: 1341–1350. http://dx.doi.org/10.1007/s13595-011-0150-7.
Ellstrand N.C., Elam D.R. (1993). Population genetic consequences of small population size: implications for plant conservation. Annual Review on Ecology, Evolution and Systematics 24: 217–242. http://dx.doi.org/10.1146/annurev.es.24.110193.001245.
Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software Structure: a simulation study. Molecular Ecology 14: 2611–2620. http://dx.doi.org/10.1111/j.1365-294X.2005.02553.x.
Excoffier L., Smouse P.E., Quattro J.M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondiral DNA restriction data. Genetics 131: 479–491.
Fady-Welterlen B. (2005). Is there really more biodiversity in Mediterranean forest ecosystems? Taxon 54: 905–910. http://dx.doi.org/10.2307/25065477.
Fady B., Lefèvre F., Vendramin G.G., Ambert A., Régnier C., Bariteau M. (2008). Genetic consequences of past climate and human impact on eastern Mediterranean Cedrus libani forests. Implications for their conservation. Conservation Genetics 8: 85–95. http://dx.doi.org/10.1007/s10592-007-9310-6.
Falk D.A., Holsinger K.E. (1991). Genetics and conservation of rare plants. Oxford University Press, New York.
Falush D., Stephens M., Pritchard J.K. (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Falush D., Stephens M., Pritchard J.K. (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Molecular Ecology Notes 7: 574–578. http://dx.doi.org/10.1111/j.1471-8286.2007.01758.x.
Farjon A. (2010). A handbook of the world’s conifers. Brill Academic Publishers, Leiden, Netherlands. http://dx.doi.org/10.1163/9789047430629.
Fennane M., Barbero M., Quézel P. (1984). Le Thuya de Berberie au Maroc: apercu phytogeographique et ecologique. Bulletin de l’Institut Scientifique de Rabat 8: 115–134.
Fischer M., Matthies D. (1998). Effects of population size on performance in the rare plant Gentianella germanica. Journal of Ecology 86: 195–204. http://dx.doi.org/10.1046/j.1365-2745.1998.00246.x.
Frankham R., Ballou J.D., Briscoe D.A. (2010). Introduction to conservation genetics. Cambridge University Press, Cambridge. http://dx.doi.org/10.1017/CBO9780511809002.
García M.S., Grau E. (2005). Aprovechamiento de los recursos le-osos en la fase protohistórica de Punta de los gavilanes (Mazarrón, Murcia). Anales de Prehistoria y Arqueología 21: 51–68.
García M.S., Grau E., Ros M.M. (2008). El Paisaje vegetal pre- y protohistórico de la costa de Mazarrón (Murcia) según el antracoanálisis de punta de los Gavilanes. Cuaternario y Geomorfología 22: 107–120.
Ge X-J., Zhou X-L., Li Z-C., Hsu T-W., Schaal B.A., Chiang T-Y. (2005). Low genetic diversity and significant population structuring in the relict Amentotaxus argotaenia complex (Taxaceae) based on ISSR fingerprinting. Journal of Plant Research 118: 415–422. http://dx.doi.org/10.1007/s10265-005-0235-1.
Grosberg R.K., Levitan D.R., Cameron B. (1996). Characterization of genetic structure and genealogies using RAPD-PCR markers: a random primer for the novice and nervous. In: Ferraris J, Palumbi SR (eds.). Molecular zoology. Advances, strategies, and protocols. Wiley-Liss, New York. p. 67–100.
Hamrick J.L., Godt M.J.W., Sherman-Broyles S.L. (1992). Factors influencing levels of genetic diversity in woody plant species. New Forests 6: 95–124. http://dx.doi.org/10.1007/BF00120641.
Jiménez J.F., Werner O., Sánchez-Gómez P., Fernández S., Guerra J. (2003). Genetic variations and migration pathway of Juniperus thurifera L. (Cupressaceae) in the western Mediterranean region. Israel Journal of Plant Sciences 51: 11–22. http://dx.doi.org/10.1560/ABR5-A6MP-5XEG-V0WF.
Kijas J.M.H., Fowler J.C.S., Thomas M.R. (1995). An evaluation of sequence tagged microsatellite site markers for genetic analysis within Citrus and related species. Genome 38: 349–355. http://dx.doi.org/10.1139/g95-045.
Levinson G., Gutman G.A. (1987). Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Molecular Biology and Evolution 4: 203–221.
Lynch M., Milligan B.G. (1994). Analysis of population genetic structure with RAPD markers. Molecular Ecology 3: 91–99. http://dx.doi.org/10.1111/j.1365-294X.1994.tb00109.x.
Magri D., Fineschi S., Bellarosa R., Buonamici A., Sebastiani F., Schirone B., Simeone M.C., Vendramin G. (2007). The distribution of Quercus suber chloroplast haplotypes matches the palaeogeographical history of the western Mediterranean. Molecular Ecology 16: 5259–5266. http://dx.doi.org/10.1111/j.1365-294X.2007.03587.x.
Médail F., Diadema K. (2009). Glacial refugia influence plant diversity patterns in the Mediterranean Basin. Journal of Biogeography 36: 1333–1345. http://dx.doi.org/10.1111/j.1365-2699.2008.02051.x.
Nei M. (1972). Genetic distance between populations. American Naturalist 106: 283–292. http://dx.doi.org/10.1086/282771.
Nei M. (1973). Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences USA 70: 3321–3323. http://dx.doi.org/10.1073/pnas.70.12.3321.
Nybom H. (2004). Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13: 1143–1155. http://dx.doi.org/10.1111/j.1365-294X.2004.02141.x.
Nybom H., Bartish I. (2000). Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspectives in Plant Ecology, Evolution and Systematics 3: 93–114. http://dx.doi.org/10.1078/1433-8319-00006.
Pérez-Collazos E., Sánchez-Gómez P., Jiménez J.F., Catalán P. (2010). The phylogeographical history of the Iberian steppe plant Ferula loscosii (Apiaceae): a test of the abundant-centre hypothesis. Molecular Ecology 18: 848–861. http://dx.doi.org/10.1111/j.1365-294X.2008.04060.x.
Petit R.M., Hampe A. (2006). Some evolutionary consequences of being a tree. Annual Review of Ecology, Evolution and Systematics 37: 187–214. http://dx.doi.org/10.1146/annurev.ecolsys.37.091305.110215.
Powell W., Morgante M, Andre C., Hanafey M., Vogel J., Tingey S., Rafalski A. (1996). The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Molecular Breeding 2: 225–238. http://dx.doi.org/10.1007/BF00564200.
Pritchard J.K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945- 959.
Pritchard J.K., Wen W. (2004). Documentation for structure software: version 2. University of Chicago, Chicago, Illinois.
Quézel P. (1980). Biogéographie et écologie des coniféres sur le pourtour méditerranéen in actualités d’écologie forestière. Gauthier-Villars, Paris.
Quézel P., Médail F. (2003). Ecologie et biogéographie des forêts du bassin méditerranéen. Elsevier, Paris.
Reed D.H., Frankham R. (2003). Correlation between fitness and genetic diversity. Conservation Biology 17: 230–237. http://dx.doi.org/10.1046/j.1523-1739.2003.01236.x.
Rodríguez-Sánchez F., Pérez-Barrales R., Ojeda F., Vargas P., Arroyo J. (2008). The Strait of Gibraltar as a melting pot for plant biodiversity. Quaternary Science Reviews 27: 2100–2117. http://dx.doi.org/10.1016/j.quascirev.2008.08.006.
Rosenbaum G., Lister G.S., Duboz C. (2002). Reconstruction of the tectonic evolution of the western Mediterranean since the Oligocene. Journal of Virtual Explorer 8: 107–130.
Sánchez-Gómez P., Stevens D., Fennane M., Gardner M., Thomas P. (2011). Tetraclinis articulata. In: IUCN 2012. IUCN red list of threatened species. Version 2012.2
Schaal B.A., Hayworth D.A., Olsen K.M., Rauscher J.T., Smith W.A. (1998). Phylogeographic studies in plants: problems and prospects. Molecular Ecology 7: 465–474. http://dx.doi.org/10.1046/j.1365-294x.1998.00318.x.
Schneider S., Roessli D., Excoffier L. (2000). Arlequin version 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland.
Slatkin M., Barton N.H. (1989). A comparison of three indirect methods for estimating average levels of gene flow. Evolution 43: 1349–1368. http://dx.doi.org/10.2307/2409452.
Sneath P.H.A., Sokal P.R. (1973). Numerical taxonomy. W.H. Freeman, San Francisco
Sytsma K.J., Givnish T.J., Smith J.F., Hain W.J. (1993). Collection and storage of land plant samples for macromolecular comparisons. Methods in Enzymology 224: 23–37. http://dx.doi.org/10.1016/0076-6879(93)24003-D.
Tautz D., Renz M. (1984). Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Research 12: 4127–4138. http://dx.doi.org/10.1093/nar/12.10.4127.
Terrab A., Paun O., Talavera S., Tremetsberger K., Arista M., Stuessy T.F. (2006). Genetic diversity and population structure in natural populations of Moroccan Atlas cedar (Cedrus atlantica; Pinaceae) determined with cpSSR markers. American Journal of Botany 93: 1114–1121. http://dx.doi.org/10.3732/ajb.93.9.1274.
Terrab A., Talavera S., Arista M., Paun O., Stuessy T.F., Tremetsberger K. (2007). Genetic diversity and geographic structure at chloroplast microsatellites (cpSSRs) in endangered West Mediterranean firs (Abies spp., Pinaceae). Taxon 56: 409–416.
Terrab A., Schönswetter P., Talavera S., Vela E., Stuessy T.F. (2008). Range-wide phylogeography of Juniperus thurifera L., a presumptive keystone species of western Mediterranean vegetation during cold stages of the Pleistocene. Molecular Phylogenetics and Evolution 48: 94–102. http://dx.doi.org/10.1016/j.ympev.2008.03.018.
Xiao L-Q., Ge X-J., Gong X., Hao G., Zheng S-X. (2004). ISSR Variation in the endemic and endangered plant Cycas guizhouensis (Cycadaceae). Annals of Botany 94: 133–138 http://dx.doi.org/10.1093/aob/mch119.
Yeh F.C., Yang R., Boyle T. (1999). POPGENE, Version 1.32. Microsoft window-based freeware for population genetic analysis. University of Alberta, Edmonton, Canada.
Young A., Boyle T., Brown T. (1996). The population genetic consequences of habitat fragmentation for plants. Trends in Ecology and Evolution 11: 413–418. http://dx.doi.org/10.1016/0169-5347(96)10045-8.
Zietkiewicz E., Rafalski A., Labuda D. (1994). Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20: 176–183. http://dx.doi.org/10.1006/geno.1994.1151.
Total of 62 references

