Chloroplast DNA polymorphism and morphometric characteristics of Carpinus betulus in the Lithuania forests
Jurkšienė G., Baliuckas V., Naugžemys D., Žvingila D. (2022). Chloroplast DNA polymorphism and morphometric characteristics of Carpinus betulus in the Lithuania forests. Silva Fennica vol. 56 no. 3 article id 10765. https://doi.org/10.14214/sf.10765
Highlights
- A 24 bp deletion was found in the chloroplast DNA region of two populations in the southeastern part of Lithuania
- Morphometric differences in hornbeam involucre between the study populations were significant
- The existence of two haplotypes of the chloroplast DNA region supports the hypothesis of two migration refugia in Carpinus betulus populations.
Abstract
The European hornbeam (Carpinus betulus L.) is a medium-sized deciduous tree that spreads northeast of the middle of Lithuania. Carpinus betulus L. is a native tree in Poland, and its branch is migrated by two Pleistocene refugia. We hypothesised that its branches had spread to Lithuania. In this study, we selected 10 populations of hornbeam that were chosen from their distribution location. We sequenced the chloroplast intergenic spacer psbA-trnH of 70 individuals. We found 24 bp deletion in chloroplast DNA (cpDNA) individuals of two populations in the southeastern part of Lithuania. In the seven forest populations, we examined the morphological variability of hornbeam seed involucres and nuts variations of 30 morphometric characteristics. Initial genetic population studies were conducted over a wider area; when differences were detected, morphological studies were conducted in the contact zone. Morphometric differences between the study populations were significant. The existence of two haplotypes of cpDNA supports the hypothesis of two migration refugia in C. betulus populations. This study contributes to significant novel knowledge about the morphological and cpDNA variability of European hornbeam populations in Lithuania and Europe.
Keywords
polymorphism;
european hornbeam;
intergenic spacer;
involucres;
migration refugia
-
Jurkšienė,
Institute of forestry, Lithuanian Research Centre for Agriculture and Forestry, Liepų str. 1 Girionys, LT-53101 Kaunas, Lithuania
 https://orcid.org/0000-0001-8210-6711
E-mail
girmante.jurksiene@lammc.lt
https://orcid.org/0000-0001-8210-6711
E-mail
girmante.jurksiene@lammc.lt

- Baliuckas, Institute of forestry, Lithuanian Research Centre for Agriculture and Forestry, Liepų str. 1 Girionys, LT-53101 Kaunas, Lithuania; Faculty of Forest Sciences and Ecology, Agriculture Academy, Vytautas Magnus University, K. Donelaičio g. 58, LT-44248 Kaunas, Lithuania E-mail virgilijus.baliuckas@lammc.lt
-
Naugžemys,
Botanical Garden of Vilnius University, Vilnius University, Kairėnų Str. 43, Vilnius 10239, Lithuania
 https://orcid.org/0000-0001-6744-5360
E-mail
genetikas@gmail.com
https://orcid.org/0000-0001-6744-5360
E-mail
genetikas@gmail.com
-
Žvingila,
Department of Botany and Genetics, Institute of Biosciences, Life Sciences Center, Vilnius University, Saulėtekio Av. 7, LT-10257 Vilnius, Lithuania
 https://orcid.org/0000-0001-7826-1815
E-mail
donatas.zvingila@gf.vu.lt
https://orcid.org/0000-0001-7826-1815
E-mail
donatas.zvingila@gf.vu.lt
Received 22 June 2022 Accepted 9 November 2022 Published 1 December 2022
Views 41101
Available at https://doi.org/10.14214/sf.10765 | Download PDF

Supplementary Files
1 Introduction
The European hornbeam (Carpinus betulus L.) (hereinafter “hornbeam”) extends from the Iberian West to the Caucasus and Northern Eastern Iran. This species is also widespread in the Balkans and the Carpathian Mountains, reaching southern Sweden in the north (Jalas and Suominen 1988, p. 62; Sikkema et al. 2016). Hornbeam has been marked as a successful tree in the north-eastern boundary through the middle of Lithuania (Navasaitis et al. 2003; Sikkema et al. 2016). Most of the late ice age and early Holocene hornbeam sediments (before 9000 BP) have been found in Italy and the eastern and south-eastern regions of the European continent (Huntley and Birks 1983). In the later periods, hornbeam spread and recolonised to northern Europe from Spain to southern Sweden (Grivet and Petit 2003) and from westwards to southern Bulgaria (Diaconeasa and Farcas 2002), Romanian Carpathians (Tantau et al. 2009), and Poland (Ralska-Jasiewiczowa et al. 2004; Granoszewski and Nalepka 2013). Isopollen maps showed that these species migrated to Poland from two different directions – southeast and west (Ralska-Jasiewiczowa et al. 2004). Boratyński et al. (2007) confirmed two different directions of hornbeam migration in Poland, based on a variation of the morphological characteristics of the nut involucres. A similar study (Legay 1989) conducted in France showed that several different refugia were identified (Legay 1989). In Lithuania, the hornbeam spread from the west and southeast in the Late Atlantic, although a small peak was observed only in the Early Subatlantic. Hornbeam was the most common in southwestern Lithuania (Kabailienė 2006).
Another study, exploring the morphological characteristics of nuts involucres and nuts, confirmed that these traits could divide hornbeam into different phenotypes (Akhondnezhad et al. 2010). However, other morphological studies on hornbeam leaves have shown that differences among hornbeam leaf characteristics depend on ecological conditions and are not taxonomically significant among the genus species (Chaplagh Paredary et al. 2012). These ecological conditions are related to ambient temperature, precipitation and altitude (Chapolagh Paridari et al. 2012). Akhondnezhad et al. (2010) concluded that environmental factors did not influence leaf length and nut weight. These leaf lengths and nut weights may be suitable for taxonomic identification of hornbeam species. However, other studies on hornbeam leaves and seeds have suggested that the physical parameters of nuts depended on the local habitat (Białobrzeska 1966; Kaliniewicz et al. 2015).
Chloroplast DNA (cpDNA) is also used to study tree demographic history and migration directions (Abbott et al. 2000; Ito et al. 2000; Mummenhoff et al. 2004; Dzialuk et al. 2009, 2017; Xiang et al. 2015; Danusevičius et al. 2021). The chloroplast intergenic spacer psbA-trnH is one of the most important variables in the genome of gymnosperm and angiosperm chloroplasts. It is also used for plant barcodes and widely used in population genetics for revealing phylogeographical patterns and population dynamics (Štorchová and Olson 2007; Hao et al. 2010; Anabat et al. 2020; Intharuksa et al. 2020; Kholina et al. 2020, 2022; Feng et al. 2022; Olsson et al. 2022). Non-coding DNA changes more rapidly than coding DNA, and mutations accumulate in it because it is not affected by selection. This property is used to study intraspecific polymorphism in plants and plant populations (Štorchová and Olson 2007; Abeysinghe 2009; Hao et al. 2010; Scarcelli et al. 2011; Luo et al. 2021). The size of the spacer psbA-trnH in various plant taxa ranges from 200 bp to 1077 bp, usually from 200 bp to 500 bp.
The study began as a part of the National Science Program “Ecosystems in Lithuania: Climate Change and Human Impact”. The aim of the project was to determine the vulnerability of the main tree species, the change in their natural ranges and the impact on biodiversity, the structure and functioning of forest ecosystems and to make forecasts (models) of change in the conditions of global changes. Therefore, the aim of this study was to investigate the polymorphism of the psbA-trnH region in hornbeam populations in Lithuania and examine whether the separated populations differ genetically and morphologically. We hypothesised that hornbeam in Lithuania comes from two migration refugia as in neighbouring Poland.
2 Materials and methods
2.1 Study population description
Lithuania is located in the northern part of the temperate zone, with a mean annual precipitation of 695 mm and a mean annual temperature of –1.1 °C in January, the coldest month. The warmest month recorded a temperature of 18.3 °C in July (LHMT 2021). Hornbeam trees are widespread in mesoeutrophic and eutrophic soils with normal moisture. They also grow on mesoeutrophic and eutrophic gleyic soils of temporarily overmoisture (Vaičys 2006). Generally, higher annual temperatures and more precipitation are recorded in the western part of Lithuania. Hornbeams are grown from the western to the southeastern part of Lithuania. The hornbeams boundary extends through the middle of the country. In the study populations, hornbeams are grown in the second forest layer with dominant species, including silver birch (Betula pendula Roth) (2, 5, 7, 8, 15 populations, Table 1; Supplementary file S1), common oak (Quercus robur L.) (3 population), common aspen (Populus tremula L.) (14 population), Scots pine (Pinus sylvestris L.) (11 population), small-leaved linden (Tilia cordata Mill.) (9, 12 populations), black alder (Alnus glutinosa (L.) Gaertn.) (1 population), or pure stand species (4, 6, 10, 13 populations).
| Table 1. Descriptive data of Carpinus betulus L. collection populations in Lithuania. | |||||||
| No | Regional Subdivision (RS), Enterprise | Geographic coordinates WGS-84 | Altitude (m) | Material (L – leaves, I – involucres, N – nuts) | Number of samples used in cpDNA analysis | Number of samples used in the morphometric analysis (I, N) | |
| X | Y | ||||||
| 1 | Kretinga RS, Mikoliškės | 2.1744353 | 69.3761656 | 70 | L | 6 | |
| 2 | Raseiniai RS Kražiai | 2.6685059 | 69.1799559 | 169 | L | 8 | |
| 3 | Tauragė RS, Pagramantis | 2.4468469 | 69.0764535 | 70 | L | 6 | |
| 4 | Radviliškis RS, Pašušvys | 3.0131742 | 69.1258463 | 107 | L | 8 | |
| 5 | Ukmergė RS, Taujėnai | 3.4118826 | 69.1347784 | 82 | L | 8 | |
| 6 | Prienai RS, Dušnionys | 3.3560720 | 68.4042971 | 191 | L, I, N | 6 | 20, 20 |
| 7 | Šalčininkai RS, Poškonys | 3.9409301 | 68.3890851 | 243 | L | 6 | |
| 8 | Prienai RS, N. Ūta | 3.1257050 | 68.5163168 | 104 | L, I, N | 6 | 20, 20 |
| 9 | Kazlų Rūda RS, Vilkaviškis | 2.7348419 | 68.5435229 | 86 | L | 6 | |
| 10 | Trakai RS Žiežmariai | 3.4003595 | 68.6523443 | 164 | L | 8 | |
| 11 | Dubrava RS, Vilkija | 2.9995270 | 68.8654302 | 30 | I, N | 20, 20 | |
| 12 | Trakai RS Būda | 3.3519082 | 68.7721338 | 86 | I, N | 20, 20 | |
| 13 | Trakai RS, Aukštadvaris | 4.8355819 | 24.5672035 | 165 | I, N | 20, 20 | |
| 14 | Varėna RS, Žygmantiškės | 3.6536445 | 68.3495198 | 150 | I, N | 20, 20 | |
| 15 | Varėna RS, Eišiškės | 3.6448808 | 68.3294573 | 156 | I, N | 20, 20 | |
2.2 DNA extraction and cpDNA analysis
For the DNA study, we selected 10 populations of hornbeam evenly distributed in the hornbeam area in different regions of Lithuania (Table 1; Suppl. file S1). Genomic DNA was extracted from the fresh leaves of 70 individuals through the modified CTAB DNA extraction method (Doyle and Doyle 1990). Amplification of the psbA-trnH region was performed as described by Shaw et al. (2005) with some modifications (Vyšniauskienė et al. 2015). The Polymerase Chain Reactions (PCR) analysis of the psbA-trnH cpDNA region was conducted in 50-μl volumes with the following reaction components: 50 ng genomic DNA, 1x Taq buffer with (NH)4SO4 (Thermo Fisher Scientific Baltics), 3 mM MgCl2, 0.2 mM each dNTP, 1.0 U Taq DNA polymerase (Thermo Fisher Scientific Baltics) and 0.2 μM each amplification primer. The PCR analysis followed these procedures: initial denaturation of 5 min at 80 °C, followed by 35 cycles of 30 s denaturation at 94 °C, 30 s annealing at 56 °C and 60 s extension at 72 °C; the final extension step was at 72 °C for 10 min. The results of the DNA fragments for the psbA-trnH region were purified from 0.8% agarose gel using a GeneJET (TM) (Enzyme Thermo Fisher Scientific) gel extraction kit. The purified DNA fragments were sequenced at the Sequencing Center of Vilnius University Life Sciences Center (Lithuania) using a 3130xl Genetic Analyzer (Applied Biosystems, USA) and a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). Amplified DNA fragments were sequenced from both ends. Sequence results were analysed using Chromas Lite 2.0 (Informer Technologies, Inc. 2008) and Mega 11 software (Tamura et al. 2021). The computer program DnaSP Version 6.12 (Rozas et al. 2017) was used to estimate InDel haplotype diversity, nucleotide diversity (Pi) and InDel diversity per site.
2.3 Morphometric measurements
Based on Boratyński et al. (2007) and our DNA analysis, we collected hornbeams involucres with nuts in locations corresponding to two possible directions of C. betulus Holocene migration from the south-east to the south-west (Table 1; Suppl. file S1). In 7 populations, nuts with involucres were cut from trees in autumn, 2021. Five trees were selected randomly, and four infructescences were collected from each tree in each population. For morphometric analysis, one involucre and one nut were taken from the middle of the infructescence. A total of 140 involucres and 140 nuts were used for the analysis. First, we measured 15 characteristics of involucres with WinFolia (Regent instrument Canada Inc. 2016) and 4 nut parameters with the calliper tool (Table 2), comprising 4 counted and 8 calculated characteristics. Nut parameters were analysed as shown in Table 2, and all involucres were analysed following the procedures developed by Boratyński et al. (2007, p. 105) (Table 2).
| Table 2. Average values and standard deviation (Mean ± SD) of characteristics, MANOVA test and F statistics of Carpinus betulus of morphometric measurements involucres and nuts in Lithuania. Numbers in bold and different letters in the same row indicate significant differences at p < 0.05. | ||||||||
| No | Morphometric traits | Abbreviations | Units | Mean ± SD | F value | p | ||
| Site 1 | Site 2 | Site 3 | ||||||
| Measured parameters with WinFolia (2016) | ||||||||
| Involucres | ||||||||
| 1 | Length | L | cm | 3.820 ± 0.63 | 3.90 ± 4.67 | 3.96 ± 0.41 | 0.15 | 0.8577 |
| 2 | Distance between top and outer base of the central lobe | DO | cm | 2.71 ± 0.46 | 2.87 ± 0.31 | 2.98 ± 0.30 | 1.07 | 0.3555 |
| 3 | Distance between top and inner base of the central lobe | DI | cm | 2.99 ± 0.53 | 3.17 ± 0.35 | 3.26 ± 0.35 | 1.03 | 0.3698 |
| 4 | Width of the central lobe at the base | WC | cm | 1.02 ± 0.11 | 1.03 ± 0.13 | 1.06 ± 0.11 | 0.26 | 0.7734 |
| 5 | Angle of top of the central lobe | AC | ° | 102.01 ± 15.23 | 97.46 ± 16.61 | 112.15 ± 16.05 | 1.6 | 0.2167 |
| 6 | Angle between central and outer lobes | ACO | ° | 48.56 ± 5.35 | 51.02 ± 6.37 | 55.09 ± 9.46 | 2 | 0.1522 |
| 7 | Length of outer lobe | LO | cm | 1.98 ± 0.42 | 1.85 ± 0.26 | 1.76 ± 0.28 | 0.95 | 0.3958 |
| 8 | Distance between top and base of outer lobe | DO | cm | 0.75 ± 0.19 | 0.71 ± 0.09 | 0.70 ± 0.18 | 0.43 | 0.6539 |
| 9 | Width of the outer lobe at the base | WO | cm | 0.60 ± 0.08 | 0.58 ± 0.08 | 0.58 ± 0.05 | 0.3 | 0.7427 |
| 10 | Angle of top of outer lobe | AO | ° | 77.39 ± 15.09 | 76.50 ± 11.08 | 78.61 ± 13.44 | 0.05 | 0.9503 |
| 11 | Angle between central and inner lobes | ACI | ° | 49.27 ± 7.49 | 54.64 ± 6.48 | 57.38 ± 12.47 | 2.74 | 0.0794 |
| 12 | Length of the inner lobe | LI | cm | 1.82 ± 0.41 | 1.73 ± 0.27 | 1.69 ± 0.17 | 0.43 | 0.6523 |
| 13 | Distance between top and base of the inner lobe | DI | cm | 0.85 ± 0.25 | 0.81 ± 0.16 | 0.81 ± 0.10 | 0.17 | 0.841 |
| 14 | Width of the inner lobe at base | WI | cm | 0.53 ± 0.06 | 0.53 ± 0.06 | 0.53 ± 0.08 | 0.07 | 0.928 |
| 15 | Angle of top of the inner lobe | AI | ° | 68.43 ± 10.52 | 73.19 ± 11.20 | 76.02 ± 12.95 | 1.15 | 0.3291 |
| Other measurements | ||||||||
| Involucres | ||||||||
| 16 | Number of teeth of the central lobe | NC | psc. | 1.92 ± 1.24 b | 5.20 ± 4.31 a | 2.10 ± 0.86 b | 5.07 | 0.0122 |
| 17 | Number of teeth of outer lobe | NO | psc. | 0.40 ± 0.46 b | 1.17 ± 0.84 a | 0.35 ± 0.29 b | 6.35 | 0.0048 |
| 18 | Number of teeth of the inner lobe | NI | psc. | 0.02 ± 0.06 b | 0.45 ± 0.48 a | 0.20 ± 0.21 ab | 6.44 | 0.0044 |
| Nuts | ||||||||
| 19 | Length | SL | mm | 7.02 ± 0.72 | 7.30 ± 0.50 | 7.02 ± 0.68 | 0.88 | 0.4255 |
| 20 | Width | SW | mm | 5.53 ± 1.64 | 5.78 ± 0.57 | 6.07 ± 0.46 | 0.45 | 0.6415 |
| 21 | Thickness | ST | mm | 3.09 ± 0.22 | 3.25 ± 0.37 | 3.29 ± 0.40 | 1.23 | 0.3066 |
| 22 | Number of edges | SE | psc. | 8.21 ± 1.34 | 8.87 ± 2.02 | 9.15 ± 1.73 | 0.692 | 0.768 |
| Counted characteristics | ||||||||
| 23 | Involucre shape of central lobe (1/4) | L/WC | 3.77 ± 0.39 | 3.82 ± 0.46 | 3.79 ± 0.67 | 0.05 | 0.9558 | |
| 24 | Involucre shape of outer lobe (7/9) | LO/WO | 3.32 ± 0.48 | 3.40 ± 0.98 | 3.21 ± 0.60 | 0.14 | 0.8711 | |
| 25 | Involucre shape of inner lobe (12/14) | LI/WI | 3.45 ± 0.62 | 3.32 ± 0.38 | 3.30 ± 0.66 | 0.3 | 0.7453 | |
| 26 | Involucre outer lobe proportion (1/7) | L/LO | 1.96 ± 0.24 b | 2.12 ± 0.16 ab | 2.29 ± 0.30 a | 4.78 | 0.0153 | |
| 27 | Involucre inner lobe proportion (1/12) | L/LI | 2.15 ± 0.27 | 2.29 ± 0.20 | 2.37 ± 0.29 | 2.07 | 0.1432 | |
| 28 | Involucre side lobe asymmetry (7/12) | LO/LI | 1.10 ± 0.08 | 1.09 ± 0.11 | 1.04 ± 0.09 | 0.61 | 0.552 | |
| 29 | Involucre asymmetry of side lobe position (2/3) | DO/DI | 0.91 ± 0.03 | 0.91 ± 0.02 | 0.91 ± 0.03 | 0.18 | 0.8391 | |
| 30 | Ratio of nut length and width (19/20) | SL/SW | 1.19 ± 0.12 | 1.28 ± 0.13 | 1.16 ± 0.05 | 2.87 | 0.0715 | |
2.4 Statistical analysis of morphometric data
Based on the DNA analysis, we divided the populations into three sites: the western site (No 8, 11, 12 in Table 1), the south-eastern site (No 6, 14, 15 in Table 1), and the intermediate site (No 13 in Table 1). We used the average of the involucres measurements and nuts of each tree, which was 35 for the calculation. We used the SAS 9.4 program (2002–2012 by SAS Institute Inc., Cary, NC, USA), using the PROC CANDISC procedure for the analysis of morphometric data (class – sites of hornbeam, variables morphometric characteristics of involucres and nuts). The CANDISC procedure derives canonical variables, which are linear combinations of quantitative variables that summarize class differences in the same way that principal components summarize total variation. In this procedure MANOVA tests the equality of the mean vector between hornbeam sites. Canonical discriminant analysis determines linear combinations of quantitative variables (involucres and nuts morphometric characteristics) that maximally discriminated the distinct refugia of hornbeam.
3 Results
3.1 cpDNA analysis
From 70 samples, the outcomes of the sequenced DNA fragments revealed only two haplotypes. All populations were monomorphic with only one haplotype. The length of the lager and most frequent haplotype was 492 bp. We found the second haplotype in populations 6 and 7 (Table 1) from the Dušnionys forest district of Prienai RS and Poškonys forest district of Šalčininkai RS, respectively. This haplotype has 24 bp deletion in positions 296–319 (Fig. 1). The total number of nucleotide sites (excluding sites with gaps) was 468. From the outcomes of analysis using the computer program DnaSP v.6.12, InDel haplotype diversity was 0,288 ± 0,059 (SD), nucleotide diversity (Pi) – 0,00308, InDel diversity per site, Pi(i) was 0,00059.

Fig. 1. Fragment of cpDNA sequences showing 24 nt deletions in the second haplotype of the psbA-trnH region was revealed in individuals of two Carpinus betulus populations (6 and 7, Table 1).
3.2 Morphometric analysis
In the sampled populations, the average values of the measured characteristics are shown in Table 1. Most of the 15 involucres measured characteristics (L, DO, DI, WC, AC, ACO, AO, ACI and AI) are 2%–14% bigger at Site 3 than at Site 1 and Site 2. Five characteristics (LO, DTO, WO, LI, DTI) were 2%–20% bigger in Site 1. Nut characteristics SW, ST and SE were 1%–10% bigger in Site 3. Counted characteristics between all three sites varied from 1% to 14%. For most characteristics of involucres and nuts, MANOVA tests showed that all mean vectors were equal (p > 0.05). Despite this, we identified statistical differences in four variables: NC, NO, NI, L/LO (p < 0.05) (No 16, 17, 18, 26, Table 1). The canonical correlation was 0.904 (p = 0.903), which was equivalent to Wilks’ lambda multivariate test. Fig. 2 displayed a plot of the first two canonical variables, showing that Can1 did not discriminate among sites. Can2 discriminates between Site 3 and the other sites.
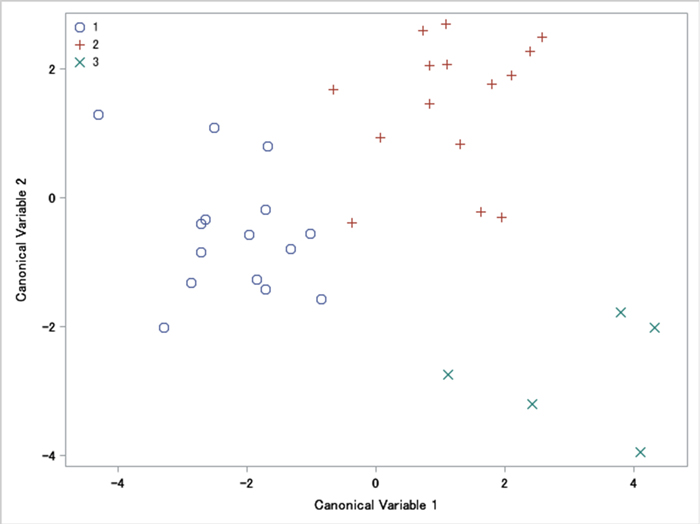
Fig. 2. A plot of the first two canonical variables of involucres and nuts morphometric traits of Carpinus betulus: 1 – western site, 2 – south-eastern site, and 3 – intermediate site. A plot of canonical variables reveals their discriminant power (in this case it is similar).
4 Discussion
This study focuses on the hornbeam, widespread in the southern part of Lithuania and occupying the northeastern edge of the hornbeam’s range. We used the psbA-trnH region to assess polymorphism in the hornbeam population. Although psbA-trnH is used to study species divergence and distinguish different species of one genus (Sang et al. 1997; Kress et al. 2005; Yoo and Wen 2007; Techaprasan et al. 2010; Yuan et al. 2010), sometimes, the cpDNA region is also used to detect intraspecific polymorphism. According to Yoo and Wen (2007), the length of the psbA-trnH intergenic spacer in Coprinus and subfamily Coryloideae ranged from 374 bp to 466 bp. Yao et al. (2009) found that the intraspecific variation among study Dendrobium species ranged from 0% to 0.1%. Jiang et al. (2017) have found that this region is suitable for intraspecific polymorphism studies. A recent study (Luo et al. 2021) has revealed the impact of environmental heterogeneity on the Acer caudatifolium Hayata population genetic diversity and demographic structure in Taiwan based on the analysis of two cpDNA regions (psbA-trnH and rpl16). Luo et al. (2021) have also found a high diversity of chlorotypes in northern Taiwan and almost monomorphic in the southern populations. Conversely, the psbA-trnH region was fixed based on the study of different cultivars of Phoenix dactylifera L. (Al-Qurainy et al. 2011) and populations of blue honeysuckle (Lonicera caerulea L.) (Naugžemys et al. 2022).
In our results, two different haplotypes were obtained in the Lithuanian hornbeam population. The 6th and 7th populations (Dušnionys and Poškonys, Suppl. file S1) were in the southeastern part of Lithuania and formed separate refugia. In Lithuania, we can distinguish two migration refugia similar to that of Poland. Jentys-Szaferova (1958, 1960) mentions fossil species of hornbeam – ancient species that appeared in Europe in morphologically differentiated form at the border of the Oligocene and Miocene. The dominant type of hornbeam in the European Tertiary period, as it is now, was a group of C. betulus. So far, all findings of this type are consistent with the current European distribution of the hornbeam. Boratyński et al. (2007) confirmed this finding after conducting research on the morphological characteristics of the nut involucres. A similar study was conducted in France, where several different refugia were identified (Legay 1989), which confirmed these two different refugia (south-eastern and western). Postolache et al. (2017) used polymerase chain reaction-restriction fragment length polymorphism and fossil pollen data for hornbeam in the Balkan Peninsula and Carpathian region, which identified three refugia: 1) the Dinaric Alps, 2) the Rhodope and Pirin Mountains and 3) the Strandzha Mountains. Through the isopollen maps, Ralska-Jasiewiczowa et al. (2004) found that the species migrated to Poland from the southeast and west. Grivet and Petit (2003) found that one major haplotype occupied western and central Europe (from Spain to southern Sweden) with all diversity restricted to south-eastern Europe.
We compared our morphometric data with the data provided by Boratyński et al. (2007), but we did not obtain significant statistical differences between the morphometric characteristics, except for No 16–18 and 26 of involucres in obtained populations. The mean values of involucres morphological characteristics of particular samples were slightly larger (No 1–4, 7–10, 12–15) than those of Boratyński et al. (2007). Three characteristics (No 5, 6 and 11) were below average. The number of teeth for two populations (1, 3) was smaller than the average, and one population (2) was larger than the maximum means. In this study, involucres with many teeth were only in the south-eastern population (Site 2), where there were differences in DNA sequence (Suppl. file S2). By the study of Jentys-Szaferova (1958) the number of teeth is a character, which is variable not only within a forest but also within an individual. Another interesting discovery by this author was that the involucres of the fossil-type hornbeam are significantly smaller than those of the present. At present, the hornbeam involucres are 3–5 cm long, and the most common length found is 3.5 cm, in our case it is 3.8–3.9 cm. The nuts of hornbeam in Miocene-Pleistocene period were smaller in 1–2 mm than in novadays (Jentys-Szaferova 1960) and the length of nuts was average in 1 mm longer in Lithuania sites than in Poland.
5 Conclusion
This study found two haplotypes of the psbA-trnH region in Lithuanian hornbeam populations, which likely indicate a different migratory history of the study populations and makes this marker potentially useful to assess the spread of hornbeam lineages. Hornbeam involucre showed significant differences in four variables (NC, NO, NI, L/LO) between study populations. The findings about the existence of polymorphism will be interesting for future research. Future studies should include DNA samples from Poland and increase the number of samples by examining the morphological characteristics of the hornbeam involucres in each population. The scientific information provides new data for illuminating the natural history of European hornbeam populations and migration in Europe.
Declaration of openness of research materials, data and code
The data are available from the authors on reasonable request.
Authors’ contributions
Conceptualization, V.B. and D.Ž.; methodology, V.B. and D.Ž.; formal analysis, G.J. and D.N.; investigation, V.B., D.Ž., D.N. and G.J.; resources, G.J. and V.B.; writing–original draft preparation, G.J. and D.Ž.; writing–review and editing, G.J. and D.Ž.; supervision, V.B. All authors have read and agreed to the published version of the manuscript.
Acknowledgements
The paper presents research findings that were obtained as part of the long-term research programme Sustainable Forestry and Global Changes implemented by the Lithuanian Research Centre for Agriculture and Forestry. We thank T. Rančelis for his valuable assistance in conducting this research.
References
Abbott RJ, Smith LC, Milne RI, Crawford RMM, Wolff K, Balfour J (2000) Molecular analysis of plant migration and refugia in the Arctic. Science 289: 1343–1346. https://doi.org/10.1126/science.289.5483.1343.
Abeysinghe P (2009) Molecular characterization of cinnamon (Cinnamomum Verum Presl) accessions and evaluation of genetic relatedness of cinnamon species in Sri Lanka based on trnL intron region, intergenic spacers between trnT-trnL, trnL-trnF, trnH -psbA and nuclear ITS. Res J Agric Biol Sci 5: 1079–1088.
Akhondnezhad S, Nejadsattari T, Sattarian A, Asri Y, Bagerieh NM (2010) The study of diversity in leaf, breack and fruit morphological characters of Carpinus betulus in various geographical conditions. J Plant Sci Res 3: 64–73.
Al-Qurainy F, Khan S, Al-Hemaid FM, Ali MA, Tarroum M, Ashraf M (2011) Assessing molecular signature for some potential date (Phoenix dactylifera L.) cultivars from Saudi Arabia, based on chloroplast DNA sequences rpoB and psbA-trnH. Int J Mol Sci 12: 6871–6880. https://doi.org/10.3390/ijms12106871.
Anabat MM, Riahi H, Sheidai M, Koohdar F (2020) Population genetic study and barcoding in Iran saffron (Crocus sativus L.). Ind Crop Prod 143, article id 111915. https://doi.org/10.1016/j.indcrop.2019.111915.
Białobrzeska M (1966) Zmienność liści i owoców grabu w analogicznych zespołach Puszczy Białowieskiej, Boreckiej i Niepołomnickiej oraz w lasach karpackich. Część I. Białowieski Park Narodowy. [Variability of the leaves and fruits of Carpinus betulus in analogous associations of the primeval forests of Puszcza Białowieska, Puszcza Borecka, Puszcza Niepołomicka and of the Carpathian Mountains. Part I. Bialowieża National Park]. Acta Soc Bot Pol 35: 401–424.
Boratyński A, Boratyńska K, Mazur M, Marcysiak K (2007) Seed involucre variation in Carpinus betulus (Corylaceae) in Poland. Acta Biol Crac Ser Bot 49: 103–111.
Chaplagh Paredary E, Jalali GA, Sunboli A, Zarafshan M (2012) Seed and bract morphotypes of Carpinus betulus. Appl Biol 25: 17–32.
Chapolagh Paridari I, Jalali SG, Sonboli A, Zarafshar M (2012) Leaf, stomata and trichome morphology of the species in Carpinus genus. TBJ 4: 11–26. https://dorl.net/dor/20.1001.1.20088906.1391.4.10.3.2.
Danusevičius D, Kembrytė R, Buchovska J, Baliuckas V, Kavaliauskas D (2021) Genetic signature of the natural gene pool of Tilia cordata Mill. in Lithuania: compound evolutionary and anthropogenic effects. Ecol Evol 11: 6260–6275. https://doi.org/10.1002/ece3.7473.
Diaconeasa B, Farcas S (2002) Aspects concernant les réfuges glaciaires, à la lumière des analyses palynologiques de séquences datées C14. [Aspects concerning glacial refuges, in the light of palynological analyzes of sequences dated C14]. Contrib Bot 37: 275–284.
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15. https://doi.org/10.2307/2419362.
Dzialuk A, Muchewicz E, Boratyński A, Montserrat JM, Boratyńska K, Burczyk J (2009) Genetic variation of Pinus uncinata (Pinaceae) in the Pyrenees determined with cpSSR markers. Plant Syst Evol 277: 197–205. https://doi.org/10.1007/s00606-008-0123-y.
Dzialuk A, Boratyńska K, Romo A, Boratyński A (2017) Taxonomic and geographic variation of the Pinus mugo complex on chloroplast microsatellite markers. Syst Biodivers 15: 464–479. https://doi.org/10.1080/14772000.2016.1257518.
Feng J, Liao F, Kong D, Ren R, Sun T (2022) Genetic diversity of the cultivated Salvia miltiorrhiza populations revealed by four intergenic spacers. Plos One 17, article id e0266536. https://doi.org/10.1371/journal.pone.0266536.
Granoszewski W, Nalepka D (2013) Carpinus betulus L.– hornbeam. In: Obidowicz A, Madeyska E, Turner C (eds) Postglacial history of vegetation in the Polish part of the Western Carpathians based on isopollen maps. W. Szafer Institute of Botany, Polish Academey of Sciences, Kraków, pp 69–76.
Grivet D, Petit RJ (2003) Chloroplast DNA phylogeography of the hornbeam in Europe: evidence for a bottleneck at the outset of postglacial colonization. Conserv Genet 4: 47–56. https://doi.org/10.1023/A:1021804009832.
Hao DC, Chen SL, Xiao PG (2010) Sequence characteristics and divergent evolution of the chloroplast psbA-trnH noncoding region in gymnosperms. J Appl Genet 51: 259–273. https://doi.org/10.1007/BF03208855.
Huntley B, Birks HJB (1983) Atlas of past and present pollen maps for Europe, 0–13,000 years ago. Cambridge University Press.
Intharuksa A, Sasaki Y, Ando H, Charoensup W, Suksathan R, Kertsawang K, Sirisa-ard P, Mikage M (2020) The combination of ITS2 and psbA-trnH region is powerful DNA barcode markers for authentication of medicinal Terminalia plants from Thailand. J Nat Med 74: 282–293. https://doi.org/10.1007/s11418-019-01365-w.
Ito M, Watanabe K, Kita Y, Kawahara T, Crawford DJ, Yahara T (2000) Phylogeny and phytogeography of Eupatorium (Eupatorieae, Asteraceae): insights from sequence data of the nrDNA ITS regions and cpDNA RFLP. J Plant Res 113: 79–89. https://doi.org/10.1007/PL00013913.
Jalas J, Suominen J (eds) (1988) Atlas Florae Europaeae: vol. 2: distribution of vascular plants in Europe. Cambridge University Press. ISBN 978-0521342711.
Jentys-Szaferowa J (1958) The Genus Carpinus in Europe in the paleobotanical literature. Monogr Bot 7: 3–59. https://doi.org/10.5586/mb.1958.005.
Jentys-Szaferowa J (1960) Morphological investigations of the fossil Carpinus nutlets from Poland. Acta Biol Crac Ser Bot 1:1–43.
Jiang D, Zhao Z, Zhang T, Zhong W, Liu Ch, Yuang Q, Huang L (2017) The chloroplast genome sequence of Scutellaria baicalensis provides insight into intraspecific and interspecific chloroplast genome diversity in Scutellaria. Genes 8, article id 227. https://doi.org/10.3390/genes8090227.
Kabailienė M (2006) Main stages of natural environmental changes in Lithuania during the Late Glacial and Holocene. Geologija 55: 37–47.
Kaliniewicz Z, Tylek P, Markowski P, Anders A, Rawa T, Liedtke M (2015) Selected physical parameters of common hornbeam (Carpinus betulus L.) nuts. Tech Sci 18: 247–259.
Kholina A, Kozyrenko M, Artyukova E, Yakubov V, Khoreva M, Andrianova E, Mochalova O, Sandanov D (2022) The species of Oxytropis DC. of section Gloeocephala bunge (Fabaceae) from Northeast Asia: genetic diversity and relationships based on sequencing of the intergenic spacers of cpDNA and ITS nrDNA. Genetica 150: 117–128. https://doi.org/10.1007/s10709-022-00152-y.
Kholina AB, Kozyrenko MM, Artyukova EV, Yakubov VV, Khoreva MG, Andrianova EA, Mochalova OA (2020) Phylogenetic relationships of Oxytropis section Arctobia of Northeast Asia according to sequencing of the intergenic spacers of chloroplast and ITS of nuclear genomes. Russ J Genet 56: 1424–1434. https://doi.org/10.1134/S1022795420120091.
Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102: 8369–8374. https://doi.org/10.1073/pnas.0503123102.
Legay JM (1989) Biometry and symmetry of hornbeam inflorescences. Can J Botany 67: 3199–3204. https://doi.org/10.1139/b89-399.
LHMT (2021) SKN - Meteo.lt. In: Lietuvos hidrometeorologijos tarnyba (LHMT). [Lithuanian Hydrometeorological Service]. http://www.meteo.lt/lt/skn. Accessed 9 Aug 2022.
Luo MX, Lu HP, Chai MW, Chai MW, Chang JT, Liao PC (2021) Environmental heterogeneity leads to spatial differences in genetic diversity and demographic structure of Acer caudatifolium. Plants 10, article id 1646. https://doi.org/10.3390/plants10081646.
Mummenhoff K, Linder P, Friesen N, Bowman JL, Lee JY, Franzke A (2004) Molecular evidence for bicontinental hybridogenous genomic constitution in Lepidium sensu stricto (Brassicaceae) species from Australia and New Zealand. Am J Bot 91(2): 254–261. https://doi.org/10.3732/ajb.91.2.254.
Naugžemys D, Patamsytė J, Žilinskaitė S, Hoshino Y, Skridaila A, Žvingila D (2022) Genetic structure of native blue honeysuckle populations in the western and eastern Eurasian Ranges. Plants 11, article id 1480. https://doi.org/10.3390/plants11111480.
Navasaitis M, Ozolinčius R, Smaliukas D, Balevičienė J (2003) Lietuvos dendroflora: monografija. [Dendroflora of Lithuania: monograph]. Lututė, Kaunas, pp 206–207.
Olsson S, Giovannelli G, Roig A, Spanu I, Vendramin GG, Fady B (2022) Chloroplast DNA barcoding genes matK and psbA-trnH are not suitable for species identification and phylogenetic analyses in closely related pines. iForest 15, article id 141.
Postolache D, Popescu F, Paule L, Ballian D, Zhelev P, Fărcaş S, Paule J, Badea O (2017) Unique postglacial evolution of the hornbeam (Carpinus betulus L.) in the Carpathians and the Balkan Peninsula revealed by chloroplast DNA. Sci Total Environ 599–600: 1493–1502. https://doi.org/10.1016/j.scitotenv.2017.05.062.
Ralska-Jasiewiczowa M, Miotk-Szpiganowicz G, Zachowicz J, Latałowa M, Nalepka D (2004) Late Glacial and Holocene history of vegetation in Poland based on isopollen maps. In: Ralska-Jasiewiczowa M. (eds) Carpinus betulus L.– hornbeam. W. Szafer Institute of Botany, Polish Academy of Sciences, Kraków, pp 69–78.
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol Biol Evol 34: 3299–3302. https://doi.org/10.1093/molbev/msx248.
Sang T, Crawford DJ, Stuessy TF (1997) Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). Am J Bot 84: 1120–1136. https://doi.org/10.2307/2446155.
Scarcelli N, Barnaud A, Eiserhardt W, Treier UA, Seveno M, dd’Anfray A, Vigouroux Y, Pintaud JC (2011) A set of 100 chloroplast DNA primer pairs to study population genetics and phylogeny in monocotyledons. Plos One 6, article id e19954. https://doi.org/10.1371/journal.pone.0019954.
Shaw J, Lickey EB, Beck JT, Farmer SB, Liu W, Miller J, Siripun KC, Winder CT, Schilling EE, Small RL (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92: 142–166. https://doi.org/10.3732/ajb.92.1.142.
Sikkema R, Caudullo G, de Rigo D (2016) Carpinus betulus in Europe: distribution, habitat, usage and threats. In: San-Miguel-Ayanz J, de Rigo D, Caudullo G, Houston Durrant T, Mauri A (eds) European atlas of forest tree species. Publ Off EU, Luxembourg, article id e01d8cf+.
Štorchová H, Olson MS (2007) The architecture of the chloroplast psbA-trnH non-coding region in angiosperms. Plant Syst Evol 268: 235–256. https://doi.org/10.1007/s00606-007-0582-6.
Tamura K, Stecher G, Kumar S (2021) MEGA 11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38: 3022–3027. https://doi.org/10.1093/molbev/msab120.
Tantau I, Reille M, de Beaulieu JL, Farcas S, Brewer S (2009) Holocene vegetation history in Romanian Subcarpathians. Quaternary Res 72: 164–173. https://doi.org/10.1016/j.yqres.2009.05.002.
Techaprasan J, Klinbunga S, Ngamriabsakul C, Jenjittikul T (2010) Genetic variation of Kaempferia (Zingiberaceae) in Thailand based on chloroplast DNA (psbA-trnH and petA-psbJ) sequences. Genet Mol Res 9: 1957–1973. https://doi.org/10.4238/vol9-4gmr873.
Vaičys M (eds) (2006) Miško augaviečių tipai. [Types of forest sites]. Lututė, Kaunas.
Vyšniauskienė R, Naugžemys D, Patamsytė J, Rančelienė V, Čėsnienė T, Žvingila D (2015) ISSR and chloroplast DNA analyses indicate frequent hybridization of alien Medicago sativa subsp. sativa and native M. sativa subsp. falcata. Plant Syst Evol 301: 2341–2350. https://doi.org/10.1007/s00606-015-1232-z.
Xiang QP, Wei R, Shao YZ, Yang ZY, Wang XQ, Zhang WC (2015) Phylogenetic relationships, possible ancient hybridization, and biogeographic history of Abies (Pinaceae) based on data from nuclear, plastid, and mitochondrial genomes. Mol Phylogenet Evol 82: 1–14. https://doi.org/10.1016/j.ympev.2014.10.008.
Yoo KO, Wen J (2007) Phylogeny of Carpinus and subfamily Coryloideae (Betulaceae) based on chloroplast and nuclear ribosomal sequence data. Plant Syst Evol 267: 25–35. https://doi.org/10.1007/s00606-007-0533-2.
Yuan WJ, Zhang WR, Han YJ, Dong MF, Shang FD (2010) Molecular phylogeny of Osmanthus (Oleaceae) based on non-coding chloroplast and nuclear ribosomal internal transcribed spacer regions. J Syst Evol 48: 482–489. https://doi.org/10.1111/j.1759-6831.2010.00099.x.
Total of 53 references.

