Genetic variation, inheritance patterns and parent–offspring relationships after artificial inoculations with Heterobasidion parviporum and Ceratocystis polonica in Norway spruce seed orchards and progeny tests
Skrøppa T., Solheim H., Steffenrem A. (2015). Genetic variation, inheritance patterns and parent–offspring relationships after artificial inoculations with Heterobasidion parviporum and Ceratocystis polonica in Norway spruce seed orchards and progeny tests. Silva Fennica vol. 49 no. 1 article id 1191. https://doi.org/10.14214/sf.1191
Highlights
- Genetic variation is demonstrated in response to artificial inoculations with Heterobasidion parviporum and Ceratocystis polonica both between parents and their offspring
- Strong relationships are observed between the male parents and their off-spring, less so between the female parents and their offspring.
Abstract
Inoculations with the two fungi Heterobasidion parviporum and Ceratocystis polonica were made in two series of progeny tests each containing full-sib families planted at two sites and on grafts of the parents in two seed orchards. Significant variation among families in lesion lengths after inoculation was found for both fungi and a predominantly additive inheritance was indicated. The estimates of narrow sense heritability were 0.13 and 0.22 for H. parviporum and C. polonica, respectively. The estimate of the genetic correlation between the lesion lengths of the two fungi was as low as 0.12. Significant variation in lesion lengths was also found among parental clones, and within ramets of the same clone, in the seed orchards. In one of the series a high positive correlation (r = 0.88) was found between the H. parviporum lesion lengths of the male parents and offspring, but not for the female parents and off-spring. The results confirm earlier conclusions that the genetic variation and heritabilities are large enough for practical breeding for resistance.
Keywords
blue stain;
heritability;
root and butt rot;
lesion length
-
Skrøppa,
Norwegian Forest and Landscape Institute, Box 115, 1431 Ås, Norway
E-mail
tore.skroppa@skogoglandskap.no

- Solheim, Norwegian Forest and Landscape Institute, Box 115, 1431 Ås, Norway E-mail halvor.solheim@skogoglandskap.no
- Steffenrem, Norwegian Forest and Landscape Institute, Box 115, 1431 Ås, Norway E-mail arne.steffenrem@skogoglandskap.no
Received 13 May 2014 Accepted 4 February 2015 Published 12 February 2015
Views 85480
Available at https://doi.org/10.14214/sf.1191 | Download PDF

1 Introduction
The main focus in practical breeding of Norway spruce (Picea abies (L.) Karst.) in the Nordic countries has up to now been on climatic adaptation, biomass production and stem and wood quality traits (Edvardsen et al. 2010). Breeding for resistance to major damaging pathogens such as the root-rot fungus (Heterobasidion parviporum Niemelä and Korhonen) and the bark beetle vectored blue-stain fungus (Ceratocystis polonica (Siem.) C. Moreau) has not been conducted, both due to limited information about genetic variation in resistance to the fungi and due to lack of reliable selection techniques. During the last 20 years several studies have demonstrated the presence of a genetic component in susceptibility to Heterobasidion, based on variation in lesion lengths after stem inoculation with the fungus (Swedjemark and Stenlid 1996, 1997; Swedjemark and Karlsson 2004; Swedjemark et al. 1997, 2001; Arnerup et al. 2010; Krokene et al. 2012; Skrøppa et al. 2014). Most of these studies were based on differently aged clonal material. Some inoculation experiments with C. polonica have also demonstrated considerable genetic variation in response to stem inoculations with this fungus both among families and among clones (Christiansen and Berryman 1995; Christiansen et al. 1999; Skrøppa et al. 2014). So far, no results from inoculations in progeny tests that are components of the practical breeding programs have been published for the two fungi and the relationships between parents and off-spring have not been studied.
In this paper results from artificial inoculations with the two fungi H. parviporum and C. polonica in two series of progeny trials with Norway spruce families from controlled crosses in two seed orchards are presented. The objectives were to characterise genetic variation in length of lesions in the inner bark after artificial inoculations of Norway spruce progenies and their parents with the two fungi and estimate genetic parameters both of this response and relationships to other growth traits and between parents and offspring.
2 Materials and methods
2.1 Experimental material and measurements
Series 1 consists of 48 full-sib families from controlled crosses between 16 grafted clones in Svenneby Seed Orchard, East Norway. This orchard produces Norway spruce seeds for altitudes between 300 and 600 m in south-eastern Norway. The parents were crossed in a factorial design to produce 48 of the 64 possible crosses, using 8 clones as mothers and 8 as fathers (Fig. 1). Two year old seedlings from all families were planted in 1990 in two short term trials on cultivated soil at spacing 1 m in a single tree plot design with 40 replicates at Hoxmark and Nilsrud, both in East Norway. The stage of flushing was scored at the beginning of growth season ten (1999) at Hoxmark on May 26 based on the method by Krutzsch (1973), and tree heights were measured at different ages in both trials. Assessments of damage such as double leaders and spike knots were made on individual trees in both trials.
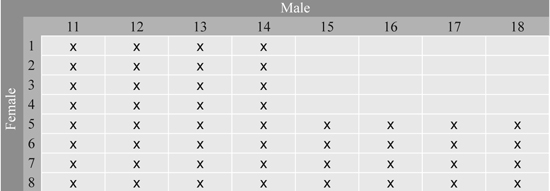
Fig. 1. Full-sib families in Series 1 from the 8 × 8 factorial cross between 16 parents from Svenneby Seed Orchard. The crosses made are denoted by x.
Series 2 consists of 20 full-sib families selected from the 100 controlled crosses in a factorial design between 10 clones used as maternal and 10 clones used as paternal parents. The selection of families was made so that each parent was involved in two crosses (Fig. 2). All parents are represented with multiple grafts in Stange Seed Orchard, Norway, where the crosses were made. This orchard produced Norway spruce seeds for altitudes below 300 m in southern Norway. Two year old seedlings from all 100 families were planted in single tree plots and with 40 replicates in two progeny trials established at forest sites in 1988, one at Skiptvedt, East Norway and the other one at Ølve, West Norway.
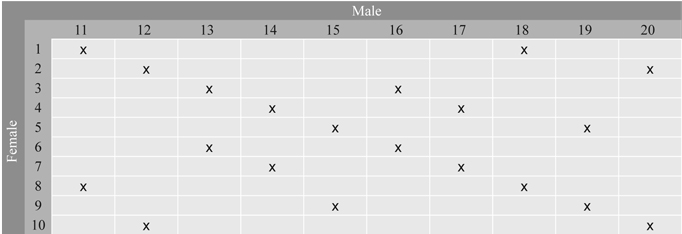
Fig. 2. Full-sib families in Series 2 selected from the 10 × 10 factorial cross between 20 parents from Stange Seed Orchard. Each parent is involved in two crosses. The families are denoted by x.
Tree heights were measured at Skiptvedt in 1997 and at Ølve in 1998. At Ølve, browsing of red deer the year before the inoculations were made resulted in considerable damage on the stem and branches. In the trial at Skiptvedt, the elongation of the terminal shoot of trees in 15 replicates of all 100 families of the factorial cross had been measured weekly during the growth season of 1992 in order to characterise the variation in timing and duration of the annual shoot growth period.
2.2 Inoculations
Inoculations with H. parviporum on young spruce trees were made near the base of the stem about 10 cm above the root collar by applying an 8-mm cork borer to remove a bark plug before an infected spruce dowel was inserted into the wound. The isolate used was H. parviporum NFLI 87-257/1. From each full-sib family the trees in 20 replicates were inoculated.
Inoculations were also made at the same time with the blue stain fungus C. polonica (Isolate NFLI 93-208/115) on the same trees. This fungus was inoculated higher up on the stem between the third and fourth branch whorl on same 20 trees in the trials. A 5-mm cork borer was used to remove a bark plug when C. polonica was inoculated. Actively growing mycelium, on malt agar (2.0% malt, 1.5% agar) was inserted in the wound before the bark plug was replaced.
Six and four ramets of each clone were inoculated at Svenneby and Stange Seed Orchards, respectively. At both sites, four inoculations were made of each of the two fungi in a circle around the stem at breast height.
All inoculations were made in the last week of June or first week of July when all trees were in active shoot growth. They were done in different years in the trials and seed orchards; in 1999 at Hoxmark and in 2000 at Nilsrud, and in 2001 at Skiptvedt and 2002 at Ølve. At Svenneby Seed Orchard inoculations were made in 2000 at Stange Seed Orchard in 2001.
The length of lesions was in all experiments measured in September, approximately 10 weeks after the inoculations were made.
2.3 Statistical methods
Plots of the residuals after preliminary analyses of variance showed deviations from normal distributions and increase in value with increasing lesion lengths. When lesion lengths were transformed by the logarithmic transformation these deviations disappeared to a large extent. Tree heights were measured after ten growth seasons at Hoxmark and after 12 growth seasons at Nilsrud and to correct for unequal variances the heights were divided by the residual standard deviation at each sites before the analyse of variance were made. The flushing scores at Hoxmark were transformed by the normal score transformation “BLOM” in the SAS RANK procedure (SAS Institute Inc. 2003SAS Institute Inc. 2003) before analyses were made.
Statistical and quantitative genetic analyses were made on the lesion lengths measured in the progeny tests and in the seed orchards and the relationships between parents and offspring were studied. Analyses were also made of tree heights and flushing scores in the trial at Hoxmark and of their relationship to the lesion lengths. A complete analysis of the shoot elongation measurements made in 1992 at Skiptvedt, based on all 100 families in the factorial, was made in Skrøppa and Steffenrem (2015), and their results for the 20 families will be used here in correlation studies.
Model (Model 1) for the statistical analyses across two sites for individual tree data in the Series 1 progeny tests:
Yhijk = μ + Sh + Fi + Mj + FMij + SFhi + SMhj + SFMhij + Bk(h) + Ehijk Here Yhijk is the observed value for the member of family ij in block k within site h, μ is the gran mean, Sh is the effect of site h, Bk(h) is the effect of block k within site h, Fi, M j and FMij are the effects of female i, male j and their interactions, respectively, SFhi, SMhj and SFMhij are their interactions with site, and Ehijk is the residual error.The 20 families tested in Series 2 can be subdivided into two groups with 10 unrelated families within each group. Separate analyses were made in each group based on the model (Model 2):
Yhijk = μ + Sh + Dij + SDhij + Bk(h) + Ehijk Here Yhijk is the observed value for the member of family ij block k within site h, μ is the grand mean, Sh is the effect of site h, Bk(h) is the effect of block k within site h, Dij is the effect of family ij, SDhij is the interaction with family and site, and Ehijk is the residual error.For the individual lesions on ramets of the grafted parents the following model (Model 3) was applied:
Yijk = μ + Ci + Rij + Eijk Here μ is the grand mean, Ci is the effect of clone i, Rij is the effect of ramet j within clone i and Eijk is the residual error for inoculation k within ramet j within clone i.In the three models, all terms, except μ and Sh, are considered to be random effects and normally and independently distributed with their respective variances.
Uni- and multivariate analysis of variance were performed on the progeny test data estimating variance components, heritabilities, genetic correlations and standard errors in a mixed model analysis in ASREML (Gilmour et al. 2009). F-tests of the variance components (Type III) were obtained from analyses in procedure GLM in SAS (SAS Institute Inc. 2003).
Genetic parameters were estimated for material I. Narrow sense heritability (h2) was estimated as 2(σ2F + σ2M) / (σ2F + σ2M + σ2FM + σ2SF + σ2SM + σ2SFM + σ2E), where σ2F is the variance estimated for the father term, σ2M is the variance for the mother term etc. from Model 1. Genetic correlations was estimated as ½(COV(x,y)F + COV(x,y)M) / √ ½(σ2xM + σ2xF)∙ ½(σ2yM + σ2yF) where COV(x,y)F is the covariance between trait x and y for the female term, COV(x,y)M is the covariance between trait x and y for the male term etc. in a multivariate model. Standard errors were estimated by the first order Taylor series expansion in ASREML (Gilmour et al. 2009).
Breeding values of the parents were estimated as BLU-predictions for the random terms of mother and father in Model 1 from the progeny test data across two sites. Pearson correlations were calculated between the parental breeding values and the mean lesion lengths of the grafts of the parents.
3 Results
3.1 Series 1
The mean lesion lengths of full-sib families after inoculation with H. parviporum varied between 23 and 49 mm in the progeny test at Hoxmark and between 21 and 37 mm in the progeny test at Nilsrud. Similar ranges of variation for C. polonica were from 31 to 78 mm and from 32 to 95 mm at Hoxmark and Nilsrud, respectively. The Pearson correlation coefficients between full-sib family means of transformed lesion lengths at the two sites were r = 0.72 and r = 0.69 for H. parviporum and C. polonica, respectively.
In the analyses of variance significant variation in lesion lengths were present among male parents for both fungi, and less so between the female parents (Table 1). The interaction between female and male parent was significant for H. parviporum (p = 0.01), but not for C. polonica. Some interaction was found between site and parents for C. polonica, but no such interactions were present for H. parviporum. The estimate of narrow sense heritability across the two sites was 0.13 for H. parviporum and 0.22 for C. polonica (Table 1).
| Table 1. Results from the analyses of variance presented by variance components and p-values for the factorial cross families at Hoxmark and Nilsrud (Series 1) for height and lesion lengths and heritabilities and type-b genetic correlations, with standard error in parentheses. Flushing was assessed at Hoxmark only. | ||||
| Source | Height | Flushing | Lesion length H. parviporum | Lesion length C. polonica |
| Site | < 0.001 | 0.18 | 0.90 | |
| Female parent | 0.036 (0.03) | 0.033 (0.0002) | 0.0011 (0.14) | 0.018 (0.03) |
| Male parent | 0.050 (0.02) | 0.213 (< 0.0001) | 0.0061 (0.0001) | 0.021 (0.003) |
| Female × male | 0.012 (0.12) | 0.019 (0.0002) | 0.0010 (0.01) | 0.004 (0.10) |
| Sites × female | 0.014 (0.03) | 0.00 (0.32) | 0.006 (0.005) | |
| Site × male | 0.020 (0.02) | 0.00 (0.23) | 0.003 (0.05) | |
| Sites × female × male | 0.027 (< 0.0001) | 0.00 (0.98)) | 0.001 (0.67) | |
| Replicate (site) | 0.158 (< 0.0001) | 0.0013 (< 0.0001) | 0.013 (< 0.0001) | |
| Error | 0.73 | 0.586 | 0.156 | 0.296 |
| Heritability | 0.20 (0.10) | 0.58 (0.20) | 0.13 (0.07) | 0.22 (0.09) |
| Type b correlation | 0.72 (0.18) | 1.00 (0.06) | 0.81 (0.13) | |
At the time of inoculation the mean tree height was 248 cm at Hoxmark and 308 cm at Nilsrud. The variance components for this trait were slightly significant for both the male and female parent, and also a significant interaction between parents and site was present (Table 1). Highly significant variation was found for the timing of flushing at Hoxmark, with an estimate of heritability h2 = 0.58.
A positive genetic correlation was present between the lesion lengths of C. polonica and the timing of flushing (Table 2), indicating longer lesions for families that had an early growth start. Between the timing of flushing and height there was a negative genetic correlation (r = –0.54 at Hoxmark) with shorter heights for the early families. This was also reflected in the negative genetic correlation between height at Hoxmark and the C. polonica lesion lengths at this site (r = –0.44), (data not shown). Taking both sites into the analysis, this correlation was strongly reduced (r = –0.19) (Table 2). The estimate of the genetic correlation between the lesion lengths of the two fungi had a low value (0.12).
| Table 2. Estimates of genetic correlations for the factorial cross families across the two sites in Series 1. | |||
| Flushing 1) | Lesion length H. parviporum | Lesion length C. polonica | |
| Height | –0.54 (0.23) | 0.08 (0.35) | –0.19 (0.34) |
| Flushing1) | 0.03 (0.40) | 0.64 (0.20) | |
| Lesion length H. parviporum | 0.12 (0.30) | ||
| 1) Flushing only assessed at Hoxmark. | |||
Significant variation in lesion lengths was also found among the grafted parental clones in the seed orchard, which had a range of variation in clonal means between 19 and 43 mm for H. parviporum (p = 0.016) and between 44 and 149 mm for C. polonica (p < 0.001) (Table 3). Significant variation in lesion lengths was also found between individual grafts (ramets) of the clones for both fungi. It is notable that the variance component for ramets within clones was of larger size than for clones for H. parviporum while it was of considerably smaller size for C. polonica (Table 3).
| Table 3. Results from the analyses of variance of lesion lengths after inoculations at Svenneby Seed Orchard (six ramets of each clones inoculated six times) (Series 1). | ||
| Source | Variance component | p-value of F-test |
| H. parviporum | ||
| Clone | 0.025 (0.016) | 0.016 |
| Ramet(clone) | 0.034 (0.013) | < 0.0001 |
| Error | 0.109 | |
| C. polonica | ||
| Clone | 0.083 (0.035) | < 0.0001 |
| Ramet(clone) | 0.031 (0.013) | 0.0001 |
| Error | 0.136 | |
For H. parviporum, a positive and significant relationship (r = 0.88) was present between the mean lesion lengths of the male parents and offspring (Fig. 3), based on estimated breeding values from the two progeny test, but not for the female parents and offspring (r = 0.16). For C. polonica, no significant correlations were found between parents and offspring (r < 0.20, p > 0.67).
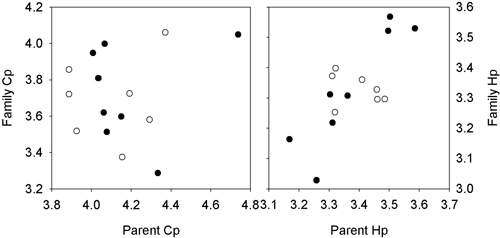
Fig. 3. Plots of the relationship between the breeding value of the female (open dots) and male parents (filled dots) against the mean value of the grafted parent for the lesions length for H. parviporum and C. polonica. Series 1: Svenneby.
3.2 Series 2
The mean lesion lengths for full-sib families after inoculation with H. parviporum varied between 29 and 62 mm at Skiptvedt and between 24 and 63 mm at Ølve. The correlation coefficient between family means at the two sites, based on the transformed values, was r = 0.55. One specific family showed extremely long lesions at both sites. Excluding this family reduced in particular the range of variation among the families at Ølve and the correlation coefficient to r = 0.25. For C. polonica, the ranges of variation among family means were between 25 and 66 mm at Skiptvedt and between 35 to 67 mm at Ølve, and with a correlation coefficient of r = 0.62 between the family means of transformed lesion lengths the two sites.
In the analyses of variance of lesion lengths across the two sites, analysed as a two independent sets of ten full-sib families, significant family differences were found in both sets for C. polonica and in one of the sets for H. parviporum (Table 4). No interactions were present between family and site. For tree heights, measured in 1997 (Skiptvedt) and 1998 (Ølve) significant family differences were present in one of the family sets.
| Table 4. Results from the analyses of variance presented by p-values for the each of the two groups of 10 independent full-sib families at Skiptvedt and Ølve (Series 2) for height and lesion lengths. | |||
| Source | Height | Lesion length H. parviporum | Lesion length C. polonica |
| Site | 0.007 0.004 | 0.01 0.0001 | 0.0006 0.38 |
| Family | 0.01 0.10 | 0.02 0.46 | 0.006 0.02 |
| Site × family | 0.l64 0.04 | 0.28 0.16 | 0.28 0.08 |
| Replicate (site) | 0.0003 0.0001 | 0.23 0.003 | 0.54 0.26 |
No relationship was found at the family level between the day of growth start and the lesion lengths for any of the two fungi (r = –0.17 and r = –0.05, data not shown). Neither were any significant relationships present between tree heights and mean lesion lengths, nor between the lesion lengths of the two fungi (data not shown).
Significant variation in lesion length was observed among the 20 clones in Stange seed orchard, with a range of variation among clones between 18 and 38 mm for H. parviporum (p < 0.001) and from 26 to 119 mm for C. polonica (p < 0.0001) (Table 5). In the analysis of the relationships between the mean lesion lengths of the families and parental means the correlation coefficients between the male parent and the off-spring were r = 0.45 (p = 0.04) and r = 0.41 (p = 0.07) for C. polonica and H. parviporum, respectively. Non-significant values were found for the relationships with the female parent (r < 0.20).
| Table 5. Results from the analyses of variance of transformed lesion lengths after inoculations at Stange Seed Orchard (four ramets of each clone inoculated four times) (Series 2). | ||
| Source | Variance component | p-value of F-test |
| H. parviporum | ||
| Clone | 0.034 (0.016) | 0.001 |
| Ramet(clone) | 0.025 (0.011) | 0.001 |
| Error | 0.102 | |
| C. polonica | ||
| Clone | 0.111 (0.043) | < 0.0001 |
| Ramet(clone) | 0.049 (0.018) | < 0.0001 |
| Error | 0.124 | |
4 Discussion
These experiments demonstrate considerable genetic variation in lesion lengths after inoculations with H. parviporum and C. polonica, both among parental clones in a seed orchard and among their progenies in trials at multiple sites. This variation is comparable with results from previous tests which in most cases have been conducted with clones propagated as rooted cuttings (Swedjemark et al. 2012). For Series 1, genetic variance components were calculated both for females and males and their interaction. These variance components were not significant in all cases, a fact that can be due to sampling effects caused by the low number of parents in each group. Generally, there was larger variation among the male than among the female parents for both fungi. The values of the female × male interaction components, although significant for H. parviporum, indicate predominantly additive inheritance, in particular for C. polonica. A good agreement was present between the responses at the two sites, even if there were some interactions between parents and sites for C. polonica. Earlier, lack of repeatability was found in experiments with clones that were inoculated under such different environmental conditions as in Sweden, Italy and Greece (Karlsson et al. 2008). The narrow sense heritability estimates of lesion lengths, h2 = 0.13 (H. parviporum) and h2 = 0.22 (C. polonica), can be compared to the quite variable broad sense heritability estimates that have been presented in other inoculation experiments (Swedjemark et al. 1997; 2001; Swedjemark and Karlsson 2004; Karlsson et al. 2008; Skrøppa et al. 2014). The values of the heritability estimates indicate a large environmental influence on these traits, a fact that is also shown by the large differences between replicates within sites and the large within family variation.
Each of the parents tested in Series 2 was involved in only two crosses. Due to this low number statistical tests were not made of individual parental effects and their interactions in the progeny tests, and no genetic parameters were estimated. Similar to Series 1, significant genetic variation in lesion lengths was found for H. parviporum in both sets of parents and in one set for C. polonica, with no interactions between families and sites. The trials Skiptvedt and Ølve in Series 2 were both planted at forest sites, while the families in Series 1 were located at cultivated soils. This may be the reason why there was a larger variability in lesion lengths within the full-sib families in Series 2 than in those in Series 1 (data not shown). At Ølve, the within-family variability in lesion lengths was largest, and family differences were smaller for H. parviporum, in particular. This could be an effect of the browsing by the red deer the winter before the inoculations were made. It has earlier been shown that mechanical wounding of the stem of Norway spruce trees may influence their resistance to C. polonica (Christiansen et al. 1999; Krokene et al. 1999). Still, there were significant relationships between the family means at the two sites for both fungi. In particular, one full-sib family had extreme lesion lengths for H. parviporum at both sites, with a mean length of 63 mm, compared to the total mean of 36 mm.
The highest heritability value (h2 = 0.58) was estimated for the flushing scores, which were assessed at the Hoxmark site only in Series 1. However, the timing of flushing for the families should be quite similar at Nilsrud, as strong relationships generally are present between flushing scores at multiple sites (Skrøppa and Steffenrem 2015). The heritability is somewhat lower than the estimates found for flushing scores in other progeny tests (e. g. Hannerz et al. 1999; Skrøppa and Steffenrem 2015). The observed genetic correlation between the timing of flushing and C. polonica lesion lengths corresponds to what was found by Krokene et al. (2012) who concluded that the level of susceptibility to this pathogen is related to tree phenology.
For height measured at the two sites Hoxmark and Nilsrud a significant interaction was found between parents and sites. This could be due to damage on leaders which had a quite high and variable frequency between families at Nilsrud, related to the timing of flushing (data not shown) and a low frequency at Hoxmark. It could also influence the estimates of genetic correlations between height and lesion lengths.
Both genetic and environmental factors contribute to the variability in lesions lengths found in the seed orchards (Tables 3 and 5). Variation in response was present both among the six or four inoculations on each stem (ramet), among the ramets within the clone and among clones. As long-living organisms trees relay on both induced and constitutive defence. Chemical compounds are thought to be important in the defence and it may vary widely between clones (Brignolas et al. 1998; Danielsson et al. 2011). However, it has earlier been shown that variation may also occur among ramets or within a single clone (Borg-Karlson et al. 1993; Persson et al. 1996) as also found here.
The relative value of the variance component for ramet within clones compared to the clonal component was larger for H. parviporum than for C. polonica. This corresponds to the higher heritability values for the last fungus found in the progeny tests. Blue-stain fungi and root rot fungi have different strategies in their living which may reflect the differences seen in this study. Pathogenic blue-stain fungi like C. polonica are introduced to the inner phloem and sapwood by the bark beetles (Christiansen and Bakke 1988; Furniss et al. 1990; Solheim 1992). For their living and further spread they are feeding on the living cells of the host tissue but also challenging the host response by the living cells. Even though H. parviporum may infect wounds (Redfern and Stenlid 1998), the main entrance is through root contacts facing the defence response in root bark (Stenlid and Redfern 1998). However, the subsequent colonization and feeding is in the lignified secondary cells of heartwood without living cells.
In Series 1, a positive relationship was demonstrated between the lesion lengths of the male parent and offspring for H. parviporum (r = 0.88), but not for C. polonica (r = 0.16). For both fungi the range of variation was wider for the male than for the female parent. In Series 2, the correlation coefficients had smaller values, and was significant only for C. polonica (r = 0.45). No significant relationships were found for the female parent and offspring. In this case, the range of variation among female parents was quite similar to that of the male parents. In an earlier study in Stange Seed Orchard, similar significant relationships were found between family means and the male parent for both fungi, and no relationship for the female parent (Skrøppa et al. 2014). As a small number of parents are involved such relationships may occur due to sampling effects. However, they may also reflect a uniparental inheritance largely controlled by the organelle genome, i. e. paternally inherited chloroplasts. A larger number of parents and families in controlled crosses, preferably with reciprocal crosses, are needed to follow up this lead.
Recently, Swedjemark et al. (2012) concluded that the genetic variation and heritabilities, estimated from inoculation studies with H. parviporum, are large enough for practical breeding purposes. The results presented here support this conclusion. However, it is necessary to investigate the relationship between actual resistance to the pathogen and the lesion length after artificial inoculation before this trait can be used as an indirect selection criterion for resistance and be included in existing breeding programs. Swedjemark et al. (2012) also stressed the need to identify new genetic markers and early traits for screening young material. For the fungus C. polonica we have shown for grafts in a seed orchard that there is strong and positive relationship between lesion lengths after single inoculations and the amount of blue stain infected wood in the stem after a mass inoculation with the same fungus (Solheim and Skrøppa, unpublished). If a similar positive relationship can be demonstrated for H. parviporum, then the response to inoculations of grafts in seed orchards or in clonal archives, or of seedlings in progeny tests, could be used as a selection criterion to select parents that have a reduced susceptibility to this fungus. The selected parents can be used to produce seed in seed orchards or to produce seedlings for the production of rooted cuttings.
Acknowledgements
We would like to thank Olaug Olsen, Anne Nilsen and Geir Østreng for technical assistance. Olaug Olsen prepared the inoculum. Anne Nilsen, Olaug Olsen and Geir Østreng did the inoculation and reading of lesion lengths. The project was financed by The Nordic Forest Research Co-operation Committee (grant SNS-77), The Research Council of Norway (grant 199346/O10), The Norwegian Forest Seed Center and The Norwegian Forest and Landscape Institute.
References
Arnerup J., Swedjemark G., Elfstrand M., Karlsson B., Stenlid J. (2010). Variation in growth of Heterobasidion parviporum in a full-sib family of Picea abies. Scandinavian Journal of Forest Research 25: 106–110. http://dx.doi.org/10.1080/02827581003730799.
Borg-Karlson A.K., Lindström M., Persson M., Norin T., Valterová I. (1993). Enantiomeric composition of monoterpene hydrocarbons in different tissues of Norway spruce Picea abies (L.) Karst. A multidimensional gas chromatography study. Acta Chemica Scandinavica 47: 138–144. http://dx.doi.org/10.3891/acta.chem.scand.47-0138.
Brignolas F., Lieutier F., Sauvard D., Christiansen E., Berryman A.A. (1998). Phenolic predictors for Norway spruce resistance to the bark beetle Ips typographus (Coleoptera: Scolytidae) and an associated fungus, Ceratocystis polonica. Canadian Journal of Forest Research 28: 720–728. http://dx.doi.org/10.1139/cjfr-28-5-720.
Christiansen E., Bakke A. (1988). The spruce bark beetle of Eurasia. In: Berryman A.A. (ed.). Dynamics of forest insect populations. Plenum Publishing Corporation. p. 479–503. http://dx.doi.org/10.1007/978-1-4899-0789-9_23.
Christiansen E., Berryman A.A. (1995). Studies of resistance in Norway spruce to bark beetle-fungus infection. In: Christiansen E. (ed.). Bark beetles, blue stain fungi, and conifer defence systems. Aktuelt fra Skogforsk 6/95. p. 26–27.
Christiansen E., Krokene P., Berryman A.A., Franceschi V.R., Krekling T., Lieutier F., Lönneborg A., Solheim H. (1999). Mechanical injury and fungal infection induce acquired resistance in Norway spruce. Tree Physiology 19: 399–403. http://dx.doi.org/10.1093/treephys/19.6.399.
Danielsson M., Lundén K., Elfstrand M., Hu J., Zhao T., Arnerup J., Ihrmark K., Swedjemark G., Borg-Karlson A.-K., Stenlid J. (2011). Chemical and transcriptional responses of Norway spruce genotypes with different susceptibility to Heterobasidion spp. infection. BMC Plant Biology 11: 154. http://dx.doi.org/10.1186/1471-2229-11-154.
Edvardsen O.M., Steffenrem A., Johnskås O.R., Johnsen Ø., Myking T., Kvaalen H. (2010). Strategi for skogplanteforedling 2010–2040 (høringsutkast). Stiftelsen Det norske Skogfrøverk / The Norwegian Forest Seed Center, Hamar.
Furniss M.M., Solheim H., Christiansen E. (1990). Transmission of blue-stain fungi by Ips typographus (Coleoptera: Scolytidae) in Norway spruce. Annals of the Entomological Society of America 83(4): 712–716. http://dx.doi.org/10.1093/aesa/83.4.712.
Gilmour A.R., Gogel B.R., Thompson R. (2009). ASReml user guide release 3.0. VSN International Ltd, Hemel Hempstead, UK.
Hannerz M., Sonesson J., Ekberg I. (1999). Genetic correlations and growth rhythm observed in short-term test and performance in long-term field trials of Norway spruce. Canadian Journal of Forest Research 29: 768–778. http://dx.doi.org/10.1139/x99-056.
Karlsson B., Swedjemark G. (2006). Genotypic variation in natural infection frequency of Heterobasidion spp. in a Picea abies clone trial in southern Sweden. Scandinavian Journal of Forest Research 21: 108–114. http://dx.doi.org/10.1080/02827580500529969.
Karlsson B., Tsopelas P., Zamponi L., Capretti P., Soulioti N., Swedjemark G. (2008). Susceptibility of Heterobasidion parviporum in Picea abies clones grown in different environments. Forest Pathology 38: 83–89. http://dx.doi.org/10.1111/j.1439-0329.2008.00543.x.
Krokene P., Christiansen E., Solheim H., Franceschi V.R., Berryman A.A. (1999). Induced resistance to pathogenic fungi in Norway spruce. Plant Physiology 12: 565–569. http://dx.doi.org/10.1104/pp.121.2.565.
Krokene P., Lahr E., Dalen L.S., Skrøppa T., Solheim H. (2012). Effect of phenology on susceptibility of Norway spruce (Picea abies) to fungal pathogens. Plant Pathology 61: 57–62. http://dx.doi.org/10.1111/j.1365-3059.2011.02487.x.
Krutzsch P. (1973). Norway spruce. Development of buds. IUFRO 2.02.11. 4 p.
Persson M., Sjödin K., Borg-Karlson A.K., Norin T., Ekberg I. (1996). Relative amounts and enantiomeric compositions of monoterpene hydrocarbons in xylem and needles of Picea abies (Pinaceae). Phytochemistry 42: 1289–1297. http://dx.doi.org/10.1016/0031-9422(96)00119-7.
Redfern D.B., Stenlid J. (1998). Spore dispersal and infection. In: Woodward S., Stenlid J., Karjalainen R., Hüttermann A. (eds.). Heterobasidion annosum. Biology, ecology, impact and control. CAB International, Oxon, New York. p. 105–124.
SAS Institute Inc. 2003. SAS/STAT user’s guide, version 9. Cary, N.C., USA.
Skrøppa T., Steffenrem A. (2015). Selection in a provenance trial of Norway spruce (Picea abies) produced a land race with desirable properties. [Submitted].
Skrøppa T., Solheim H., Hietala A.M. (2014). Variation in phloem resistance of Norway spruce clones and families to Heterobasidion parviporum and Ceratocystis polonica and its relationship to phenology and growth traits. Scandinavian Journal of Forest Research. http://dx.doi.org/10.1080/02827581.2014.963144.
Solheim H. (1992). The early stages of fungal invasion in Norway spruce infested by the bark beetle Ips typographus. Canadian Journal of Botany 70: 1–5. http://dx.doi.org/10.1139/b92-001.
Stenlid J., Redfern D.B. (1998). Spread within the tree and stand. In: Woodward S., Stenlid J., Karjalainen R., Hüttermann A. (eds.). Heterobasidion annosum. Biology, ecology, impact and control. CAB International, Oxon, New York. p. 125–141.
Swedjemark G., Karlsson B. (2004). Genotypic variation in susceptibility following artificial Heterobasidion annosum inoculation of Picea abies clones in a 17-year-old field test. Scandinavian Journal of Forest Research 19: 103–111. http://dx.doi.org/10.1080/02827580310018032.
Swedjemark G., Stenlid J. (1996). Variation in spread of Heterobasidion annosum in clones of Picea abies grown at different vegetation phases under greenhouse conditions. Scandinavian Journal of Forest Research 11: 137–144. http://dx.doi.org/10.1080/02827589609382921.
Swedjemark G., Stenlid J. (1997). Between-tree and between-isolate variation for growth of S-group Heterobasidion annosum in sapwood of Picea abies cuttings. Canadian Journal of Forest Research 27: 711–715. http://dx.doi.org/10.1139/x96-191.
Swedjemark G., Stenlid J., Karlsson B. (1997). Genetic variation among clones of Picea abies in resistance to growth of Heterobasidion annosum. Silvae Genetica 46: 369–374.
Swedjemark G., Stenlid J., Karlsson B. (2001). Variation in growth of Heterobasidion annosum among clones of Picea abies incubated for different periods of time. Forest Pathology 31: 163–175. http://dx.doi.org/10.1046/j.1439-0329.2001.00238.x.
Swedjemark G., Borg-Karlsson A.K., Karlsson B. (2012). Breeding for resistance in Norway spruce to the root and butt fungi Heterobasidion spp. USDA Forest Service, Pacific Southwest Research Station, General Technical Report PSW-GTR-240. p. 162–166.
Total of 29 references

