Defoliation of Tilia cordata trees associated with Apiognomonia errabunda infection in Finland
Vainio E. J., Velmala S. M., Salo P., Huhtinen S., Müller M. M. (2017). Defoliation of Tilia cordata trees associated with Apiognomonia errabunda infection in Finland. Silva Fennica vol. 51 no. 4 article id 7749. https://doi.org/10.14214/sf.7749
Highlights
- Defoliation of Tilia cordata was investigated by fungal isolation from symptomatic leaf petioles and ITS sequence determination
- The disease symptoms were associated with the presence of Apiognomonia errabunda
- We report the first nucleotide sequences of A. errabunda from the Nordic countries.
Abstract
We investigated the causative agent of a disease outbreak affecting small-leaved limes (Tilia cordata Mill.) and resulting in darkening of the leaf petioles and excessive defoliation during summer 2016 in southern Finland. The fungal species composition of the symptomatic petioles was examined by culture isolation and molecular identification using ITS rDNA sequences, which revealed the most prevalent fungal species present in the petioles as Apiognomonia errabunda (Roberge) Höhn. Based on reviewing curated herbarium specimens deposited at the Universities of Helsinki and Turku, A. errabunda is native and widely distributed in small-leaved limes in Finland, and occasionally infects also other broadleaved trees, including Quercus robur L. and ornamental species of Tilia L. and Fagus L. The ITS sequence analysis conducted during this study revealed minor within-species polymorphisms similar to those observed earlier in the Central European and Russian populations of A. errabunda, and reports the first nucleotide sequences of this species from the Nordic countries.
Keywords
small-leaved lime;
fungal disease;
anthracnose;
ITS rDNA;
direct PCR;
Tiliaceae
-
Vainio,
Natural Resources Institute Finland (Luke), Management and Production of Renewable Resources, P.O. Box 2, FI-00791 Helsinki, Finland
 http://orcid.org/0000-0002-6739-7968
E-mail
eeva.vainio@luke.fi
http://orcid.org/0000-0002-6739-7968
E-mail
eeva.vainio@luke.fi

- Velmala, Natural Resources Institute Finland (Luke), Management and Production of Renewable Resources, P.O. Box 2, FI-00791 Helsinki, Finland E-mail sannakajsa.velmala@luke.fi
- Salo, Finnish Museum of Natural History, Botanical Museum, P.O. Box 7, FI-00014 University of Helsinki, Finland E-mail pertti.salo@helsinki.fi
- Huhtinen, University of Turku, Herbarium, Biodiversity Unit, FI-20014 Turku, Finland E-mail sephuh@utu.fi
- Müller, Natural Resources Institute Finland (Luke), Management and Production of Renewable Resources, P.O. Box 2, FI-00791 Helsinki, Finland E-mail micms.muller@gmail.com
Received 9 June 2017 Accepted 18 August 2017 Published 23 August 2017
Views 63707
Available at https://doi.org/10.14214/sf.7749 | Download PDF

Supplementary Files
1 Introduction
Tilia cordata Mill. (small-leaved lime) is a deciduous tree distributed widely in Central Europe, but lacking from the southernmost Mediterranean region as well as northern Fennoscandia (Radoglou et al. 2009). In Finland, it reaches its northern limit below latitude 63°44´N (Lampinen and Lahti 2016; Raino Lampinen, University of Helsinki, personal comment in 2017) and has a fragmented distribution on fertile soils. In the north, small-leaved limes are capable of producing mature seeds only during warm summers, and they propagate mainly by sprouting (Pigott and Huntley 1981). T. cordata trees are also commonly planted in private gardens, whereas common limes, i.e., Tilia × vulgaris Hayne (syn. Tilia × europaea L., hybrid of Tilia cordata and Tilia platyphyllos Scop. syn. Tilia grandifolia Ehrh.) are often planted in public parks and as streetside trees.
Tilia trees are considered to be relatively resistant to pathogens, but various pathogenic fungi commonly affect T. cordata trees suffering from anthropogenic disturbances in urban environments. In central Europe, the most common leaf spot diseases include Mycosphaerella millegrana (Cooke) J. Schröt. and Apiognomonia errabunda (Roberge) Höhn. (Snieškienė et al. 2012; Stravinskienė et al. 2015), and Asteroma tiliae F. Rudolphi and Septoria tiliae Westend. are also commonly detected (Snieškienė et al. 2012). Various sooty molds, such as Aureobasidium pullulans (de Bary) G. Arnaud, Cladosporium herbarum (Pers.) Link and Leptoxyphium fumago (Woron.) R. C. Srivast. occur frequently on T. cordata leaves (Snieškienė et al. 2012). Several fungal pathogens like Nectria cinnabarina (Tode) Fr., Chondrostereum purpureum (Pers.) Pouzar and Armillaria spp. (Fr.) Staude infect Tilia branches, bark wounds and wood material.
In early June 2016, small-leaved limes at several sites in Southern Finland were reported to suffer from severe defoliation and darkening of the leaf petioles. In order to examine whether the symptoms were associated with known fungal pathogens of T. cordata, we investigated the fungal species composition of the symptomatic petioles by culture isolation and molecular identification using internal transcribed spacer (ITS) ribosomal DNA sequences. After identifying the most prevalent fungal species present in the leaf petioles as A. errabunda, the occurrence of this known leaf pathogen in Finland was reviewed based on curated museum specimens deposited at the Universities of Helsinki and Turku.
2 Material and methods
2.1 Sampling sites with Tilia cordata defoliation symptoms
Symptomatic T. cordata leaves were collected from three locations in southern and central Finland in early June, 2016: Kiikala (Salo, Varsinais-Suomi; 60°27´N, 23°35´E), Aulanko (Hämeenlinna, Kanta-Häme; 61°01´N, 24°27´E), and Akaa (Pirkanmaa; 61°07´N, 23°56´E). The sampling dates were 6 June 2016, 8 June 2016 and 8 June 2016, respectively. A second sampling was conducted at the Kiikala site in 1 August 2016. The sampling sites at Kiikala and Akaa are private gardens, and the Aulanko site is a protected forest park.
The most prominent symptoms were excessive leaf loss (Figs. 1A, C) and blackening of the leaf petioles (Fig. 1B). The Aulanko and Kiikala sites were inspected again in August, and the trees still suffered from defoliation as well as leaf anthracnose symptoms that had developed during the summer. At the Kiikala site, similar symptoms were reported already in 2015 and again in June 2017. At each sampling site, 3–12 neighboring T. cordata trees were severely affected by the disease.

Fig. 1. Disease symptoms caused by Apiognomonia errabunda. (A) Small-leaved lime trees suffering from defoliation in Aulanko, Hämeenlinna; (B) Leaves of Tilia cordata with darkened petioles collected from Kiikala, Salo; (C) leaf loss of small-leaved limes in Kiikala, Salo; (D) A. errabunda strain 3C2V representing the dominating morphotype retrieved from the leaf petioles cultured on MOS agar plate covered with a cellophane membrane. Pictures A, B and C were provided by Tapio Ykspetäjä and Seppo Lagom.
According to the weather data collected by the Finnish Meteorological Institute (http://en.ilmatieteenlaitos.fi), the mean temperature in May 2016 was 2.4–3.4 degrees above mean in the Hämeenlinna-Salo region with an average temperature of 10.1–10.4 °C in May during 1981–2010, while June temperatures were near average (<1 degrees above the mean temperature of 14.3–14.7 °C). The rainfall in June was slightly higher than normal (111–125% of mean June rainfall of 56.8–66.3 mm during 1981–2010), whereas May rainfall was 76% and 105% of average in Hämeenlinna and Salo, respectively (with mean June rainfall of 40.3 and 37.1 mm).
2.2 Direct polymerase chain reaction (PCR) from Tilia cordata leaf petioles
Small pieces of blackened leaf petioles were initially sampled with Harris Unicore™ sampling tool (Ø 1 mm) from five symptomatic T. cordata leaves (one from Kiikala and two from each of Aulanko and Akaa) and subjected to direct polymerase chain reaction (PCR) amplification with ITS primers ITS1F and ITS4 (Gardes and Bruns 1993; White et al. 1990) using the dilution protocol of Phire Plant Direct PCR Kit (Thermo Fischer Scientific) as described earlier by Velmala et al. (2013). Resulting amplicons were extracted from 1% agarose gel supplemented with synergel (Diversified Biotech) and purified with the EZNA Biotek gel extraction kit (Omega Biotek) and subjected to sequencing in both directions with primers ITS1F and ITS4 at Macrogen Inc., Korea (http://www.macrogen.com).
2.3 Fungal cultures isolated from blackened Tilia cordata leaf petioles
A second batch of leaf samples was collected in 1 August 2016 from three diseased T. cordata trees at the Kiikala site and subjected to culture isolation. Blackened leaf petioles (N = 20) were dispatched from the leaves and surface sterilized. In order to discard putative surface contaminants while preserving the viability of fungal mycelia residing within the petiole tissues, we tested five different surface sterilization protocols, each carried out using four petioles: (1) no surface sterilization, (2) 30 s incubation in 70% ethanol, (3) incubation of 30 s in 70% ethanol, 30 s in 5% NaOCl and 2× 30 s in 70% ethanol (4) incubation of 30 s in 70% ethanol, 60 s in 5% NaOCl and 2× 30 s in 70% ethanol (5) incubation of 30 s in 70% ethanol, 4 min in 5% NaOCl and 2× 30 s in 70% ethanol. The leaf petioles were then placed on malt extract agar and water agar plates (two and two petioles per treatment type) and incubated at 15 °C until the appearance of fungal hyphae. Fungal colonies emerging within three weeks of incubation were subcultured on modified orange serum (MOS) agar plates covered with cellophane membranes, incubated in ambient light at room temperature and morphotyped.
Representative isolates from each morphotype were selected for ITS sequence analysis using direct PCR with the Phire II Hot Start DNA polymerase as described above, but using a small amount of mycelia scraped directly from the membrane-covered MOS agar plates. According to the manufacturer (Thermo Fischer Scientific), the DNA polymerase used has an error rate of 1.14 × 10–5.
2.4 Internal transcribed spacer (ITS) sequence analysis
Global sequence alignments were constructed using Geneious R10 version 10.0.8 (Biomatters Ltd.) and MAFFT alignment algorithms. Sequence comparisons were conducted using NCBI BlastN (nucleotide collection) and the UNITE database (Kõljalg et al. 2013).
2.5 Herbarium collections
Herbarium specimens deposited at the Universities of Helsinki (LUOMUS) and Turku were revisited to investigate the earlier occurrences of A. errabunda in Finland. The survey included also specimens deposited using the current or past anamorph names of the fungus (e.g., Gloeosporium tiliae Oudem, Gloeosporium umbrinellum Berk. & Broome, Discula umbrinella (Berk. & Broome) M. Morelet; see MycoBank at http://www.mycobank.org and Index Fungorum at http://www.indexfungorum.org).
3 Results
3.1 Fungal morphotypes
Altogether 31 fungal isolates were retrieved from the 20 leaf petioles collected from the Kiikala sampling site (1–3 distinct colonies from each petiole; 5, 7, 6, 8 and 5 colonies per sterilization treatment protocol 1–5, respectively). The majority of these cultures (N = 25) formed light gray velutinous mycelial colonies on MOS plates, part of them with concentric dark rings or lobed colony margins (Fig. 1D). The remaining six cultures represented mostly dark grey spotted or moldlike morphotypes. Four of these moldlike cultures were obtained from petioles plated without surface sterilization.
3.2 Internal transcribed spacer (ITS) sequences analysis
Using direct PCR, we obtained amplification products of the expected size from two leaf petiole samples from Akaa and one sample from Aulanko. The corresponding high quality ITS sequences covered the entire ITS1–5.8S rDNA–ITS2 region of 501 nt. No amplification products were obtained by direct PCR from the Kiikala sample, whereas ITS sequences were successfully determined from all the 13 cultured isolates from Kiikala selected for sequence analysis.
Based on ITS sequence analysis, the most common morphotype constituting 81% of the retrieved cultures from Kiikala was identified as A. errabunda. This species constituted 20%, 86%, 100%, 88%, and 100% of the cultures obtained using surface sterilization protocols 1–5, respectively. The other morphotypes observed more than once were identified as Botrytis sp. P. Micheli, Cladosporium sp. Link and Monilinia sp. Honey (Table 1).
| Table 1. ITS sequence information. View in new window/tab. |
There were three different ITS sequence variants representing A. errabunda among the Kiikala isolates, two of which were also detected by direct PCR in Aulanko and/or Akaa (Table 1). These sequence variants were highly similar and contained only 1–2 differing nucleotide sites corresponding to 99.6–99.8% overall sequence identity. One sequence variant detected in Akaa and Kiikala showed 100% identity to those of A. errabunda isolates from T. cordata in Novgorod, Russia. Another sequence variant found at all three sampling sites shared 100% identity with the ITS sequence of A. errabunda isolated from fireweed (Chamaenerion angustifolium L. Scop.) in Switzerland (Sogonov et al. 2007). There was also one unique sequence variant with an insertion of a single G nucleotide after position 48 of the ITS2 sequence (GenBank accession KY927820). All sequences lacked the A insertion at sequence position 182 characteristic of Apiognomonia veneta (Sacc. & Speg.) Höhn. (Sogonov et al. 2007). The sequences determined in this study were deposited in GenBank under the accession numbers KY927820-KY927835.
3.3 Previous observations of Apiognomonia errabunda in Finland based on herbarium specimens
There were a total of 27 and 6 curated specimens of A. errabunda in the herbaria of the Universities of Helsinki and Turku, respectively. Detailed information is reported in Supplementary file S1, which lists the A. errabunda specimens of the University of Helsinki with corresponding URLs at the Finnish Biodiversity Information Facility (FinBIF; https://www.laji.fi). Specimens of the University of Turku are described at the Research portal Mkk Kotka (TUR, http://mus.utu.fi/TFU.62600; TUR, http://mus.utu.fi/TFU.77828; TUR, http://mus.utu.fi/TFU.107022; TUR, http://mus.utu.fi/TFU.116689; TUR, http://mus.utu.fi/TFU.153601; TUR, http://mus.utu.fi/TFU.172978). The collection sites of herbarium specimens and the leaf samples collected during this study are shown in Fig. 2.
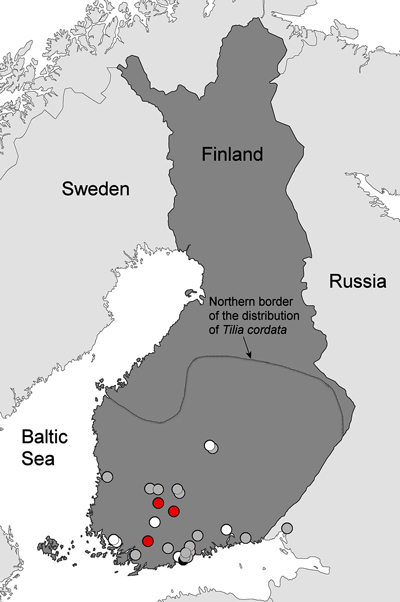
Fig. 2. Historical records of Apiognomonia errabunda in Finland and the sampled disease outbreaks on small-leaved lime in 2016. The 2016 disease outbreaks were observed in three locations (red dots). Sampling sites of A. errabunda specimens in the herbaria of University of Helsinki (LUOMUS) and University of Turku date from 1870 to 2004 and are plotted with the following symbols indicating host tree species: gray = Tilia cordata, black = Tilia × europaea or white dots = other or unknown tree species. The northern border of the distribution area of Tilia cordata in Finland is given according to Lampinen and Lahti (2016).
The oldest available museum specimen of A. errabunda in Finland dates from 1870 and was identified by P. A. Karsten as Gnomonia petiolicola (Fuckel) P. Karst. Based on re-examining this sample, Monod (1983) later identified it to be A. errabunda (Suppl. file S1). During the following decades, this species has been detected at several locations in southern and southwestern Finland, and the northernmost available samples originate from Jyväskylä (62°14´N, 25°45´E). The most common host species is T. cordata, but there are also single samples collected from T. platyphyllos, Quercus robur, Tilia × vulgaris and Fagus sylvatica L. (the latter two are non-native tree species used as ornamentals in Finland). Most samples have been named according to the anamorph state of A. errabunda, namely Gloeosporium tiliae (Suppl. file S1).
4 Discussion
In this study, we identified Apiognomonia errabunda as the most probable cause of the disease symptoms observed in small-leaved lime trees in early summer 2016. This fungal species was the most common culture morphotype isolated from blackened leaf petioles, and the only species detected using direct PCR amplification. The observed symptoms also supported A. errabunda as the causative agent: reported disease symptoms include brown leaf spots with dark margins as well as defoliation in early summer, followed by wilting and death of branches (Horst 2001). Since Karsten’s early observations (Karsten 1873), the disease has been described by its anamorph name Gloeosporium tiliae in Finnish plant pathology textbooks (Liro 1924; Rauhala 1958), which mention this species to be common. Pohjakallio (1963) describes the disease on lime and oak trees using the name Discula quercina (Cooke) Sacc. (=Gloeosporium umbrinellum). A. errabunda has a wide geographical distribution in the temperate Northern Hemisphere including central and Eastern Europe and North America, and occurs on several important forest trees, such as Quercus spp. L., Fagus sylvatica, Populus tremula L. and Castanea sativa Mill. (Sogonov et al. 2007). However, the occurrence of this fungal pathogen in Finland has not been systematically reviewed in scientific literature before this report.
Moreover, prior to this study there were no GenBank records reporting nucleotide sequences of A. errabunda from any of the Nordic countries, although it has been recorded to occur also in Sweden and Denmark (Sogonov et al. 2007; Eriksson 2014; Sundelin et al. 2015). The ITS sequence analysis conducted during this study revealed minor within-species polymorphisms similar to those observed earlier in the Central European and Russian populations of A. errabunda. Thus, one of the common sequence variants detected in this study is identical with the A. errabunda sequence described by Sogonov et al. (2007) from Novgorod, Russia. This sequence variant originates from T. cordata, and represents the taxonomical clade formerly classified as Apiognomonia tiliae (Rehm) Höhn. (Sogonov et al. 2007; see also the description by Dr. Paul Cannon from the Mycology Department at Kew (http://fungi.myspecies.info/). However, the second sequence variant observed in Finland clusters outside this “tiliae clade” of A. errabunda (Sogonov et al. 2007). It should be noted that the latter sequence was also 100% identical with unpublished ITS sequences assigned to Fusicoccum quercus Oudem. (Table 1). According to the MycoBank database (http://www.mycobank.org), F. quercus may be a synonym of Apiognomonia errabunda, and the fungus causes bark cankers on oak trees (Ragazzi et al. 2003). However, Sogonov et al. (2007) conclude that the unpublished sequences assigned as F. quercus (AY853206-14 and AJ293872-5; see Table 1) actually represent A. errabunda and not F. quercus (the anamorphic state of Botryosphaeria Ces. & De Not.). Overall, only four polymorphic sites have been reported in the ITS1-5.8S-ITS2 region of A. errabunda (Sogonov et al. 2007).
From an epidemiological point of view, it may be noteworthy that in 2016 the noticeable disease symptoms were observed soon after a massive amount of aphids drifted via southwestern winds into Finland in May 15th (Huusela-Veistola 2016). The rarer fungal morphotypes isolated from the leaf petioles during this study (Botrytis sp. and Cladosporium sp.) have been described to cause sooty mold in Tilia and are typically associated with aphid infestation (Snieškienė et al. 2012).
We conclude that the disease symptoms observed in early summer 2016 in T. cordata can be associated with the presence of A. errabunda, which is a native and widely distributed species in Finland causing occasional disease outbreaks. Future studies are needed to investigate the host range and epidemiology of A. errabunda in the Nordic countries. At the Kiikala sampling site, there were five healthy common lime trees (Tilia × vulgaris) growing next to twelve small-leaved lime trees, all of which were affected by the disease in 2015 and 2016 (Seppo Lagom, pers. comm. 2016). Moreover, the vast majority of Finnish museum specimens originated from T. cordata, and there were only single occurrences of the disease on common lime or large-leaved lime (Tilia grandifolia = T. platyphyllos). These observations suggest that our native species T. cordata may be more susceptible to A. errabunda than the hybrid limes commonly planted as park and streetside trees.
Acknowledgements
We are grateful to Sami Kiema, Tapio Ykspetäjä, Seppo Lagom and Johanna Ahtiainen for providing leaf samples and pictures for the study. Marja-Leena Santanen and Juha Puranen are acknowledged for skillful technical assistance, and Erja Huusela-Veistola, Saara Velmala and Tea von Bonsdorff are thanked for valuable scientific discussions.
References
Eriksson O. (2014). Checklist of the non-lichenized ascomycetes of Sweden. Symbolae botanicae Upsalienses 36(2). 499 p.
Gardes M., Bruns T.D. (1993). ITS primers with enhanced specificity for basidiomycetes‐application to the identification of mycorrhizae and rusts. Molecular Ecology 2(2): 113–118. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x.
Horst R.K. (2001). Westcott’s plant disease handbook. Sixth edition. Springer Science+Business Media, New York. 1032 p. https://doi.org/10.1007/978-1-4757-3376-1.
Huusela-Veistola E. (2016). Tuulen tuomaa 2016 – kaalikoita ja kirvoja kaukokulkeumana. [Migration of diamondback moths and aphids in 2016]. Kasvinsuojelulehti 49(3): 72–75.
Karsten P.A. (1873). Mycologia fennica, Pars secunda: Pyrenomycetes. [Fungi of Finland, part two: Pyrenomycetes]. Bidrag till Kännedom av Finlands Natur och Folk. 23: 1–252.
Kõljalg U., Nilsson R.H., Abarenkov K., Tedersoo L., Taylor A.F.S., Bahram M., Bates S.T., Bruns T.D., Bengtsson-Palme J., Callaghan T.M., Douglas B., Drenkhan T., Eberhardt U., Dueñas M., Grebenc T., Griffith G.W., Hartmann M., Kirk P.M., Kohout P., Larsson E., Lindahl B.D., Lücking R., Martín M.P., Matheny P.B., Nguyen N.H., Niskanen T., Oja J., Peay K.G., Peintner U., Peterson M., Põldmaa K., Saag L., Saar I., Schüßler A., Scott J.A., Senés C., Smith M.E., Suija A., Taylor D.L., Telleria M.T., Weiss M., Larsson K.-H. (2013). Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology 22(21): 5271–5277. https://doi.org/10.1111/mec.12481.
Lampinen R., Lahti T. (2016). Kasviatlas 2015. [Plant atlas 2015]. Helsingin Yliopisto, Luonnontieteellinen keskusmuseo, Helsinki. http://www.luomus.fi/kasviatlas.
Liro J.I. (1924). Tuhosienet: tärkeimmät tuhosienet metsänhoitajia, maanviljelijöitä, puutarhureja sekä kasvitiedettä opiskelevia varten. [Fungal plant pathogens]. Vanamon kirjoja n:o 22. Otava, Finland. 405 p.
Monod M. (1983). Monographie taxonomique des Gnomoniaceae. [A taxonomical monograph on Gnomoniaceae]. Beihefte zur Sydowia, Annales Mycologici, Ser. II , IX. Beiheft. 315 p.
Pigott C.D., Huntley J.P. (1981). Factors controlling the distribution of Tilia cordata at the northern limits of its geographical range III. Nature and causes of seed sterility. New Phytologist 87(4): 817–839. https://doi.org/10.1111/j.1469-8137.1981.tb01716.x.
Pohjakallio O. (1963). Kasvipatologia 2. [Plant pathology 2]. WSOY, Porvoo, Finland. 375 p.
Radoglou K., Dobrowolska D., Spyroglou G., Nicolescu V.N. (2009). A review on the ecology and silviculture of limes (Tilia cordata Mill., Tilia platyphyllos Scop. and Tilia tomentosa Moench.) in Europe. Die Bodenkultur 60: 7–17.
Ragazzi A., Moricca S., Capretti P., Dellavalle I., Turco E. (2003). Differences in composition of endophytic mycobiota in twigs and leaves of healthy and declining Quercus species in Italy. Forest Pathology 33(1): 31–38. https://doi.org/10.1046/j.1439-0329.2003.3062003.x.
Rauhala A. (1958). Kasvien sienitauteja. [Fungal plant diseases]. WSOY, Finland. 354 p.
Snieškienė V., Baležentienė L., Stankevičienė A., Meškauskienė V. (2012). Intensity of fungal diseases of small-leaved lime (Tilia cordata Mill.) across urban greeneries of Lithuania. Journal of Food, Agriculture and Environment 10: 988–993.
Sogonov M.V., Castlebury L.A., Rossman A.Y., White J.F. (2007). The type species of Apiognomonia, A. veneta, with its Discula anamorph is distinct from A. errabunda. Mycological Research 111(6): 693–709. https://doi.org/10.1016/j.mycres.2007.03.013.
Stravinskienė V., Snieškienė V., Stankevičienė A. (2015). Health condition of Tilia cordata Mill. trees growing in the urban environment. Urban Forestry & Urban Greening 14(1): 115–122. https://doi.org/10.1016/j.ufug.2014.12.006.
Sundelin T., Strømeng G.M., Gjærum H.B., Amby D.B., Ørstad K., Jensen B., Lund O.S., Stensvand A. (2015). A revision of the history of the Colletotrichum acutatum species complex in the Nordic countries based on herbarium specimens. FEMS Microbiology Letters 362(16): fnv130. https://doi.org/10.1093/femsle/fnv130.
Velmala S.M., Rajala T., Haapanen M., Taylor A.F.S., Pennanen T. (2013). Genetic host-tree effects on the ectomycorrhizal community and root characteristics of Norway spruce. Mycorrhiza 23(1): 21–33. https://doi.org/10.1007/s00572-012-0446-y.
White T.J., Bruns T., Lee S.B., Taylor J.W. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J. (eds.). PCR protocols: a guide to methods and applications. Academic Press, London. p. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1.
Total of 20 references.

