Optimizations of high throughput multiplex polymerase chain reaction with simple sequence repeat markers for genotyping of common walnut populations (Juglans regia L.)
Ćelepirović N., Karija Vlahović M., Dounavi A., Ivanković M. (2016). Optimizations of high throughput multiplex polymerase chain reaction with simple sequence repeat markers for genotyping of common walnut populations (Juglans regia L.). Silva Fennica vol. 50 no. 5 article id 1674. https://doi.org/10.14214/sf.1674
Highlights
- We combined eleven SSR markers in one multiplex PCR to make faster and cost effective amplification of the common walnut DNA from Croatia
- Genetic variation of common walnut from Croatia was moderate at analyzed SSR loci
- The resultant multiplex PCR could be used for genotyping of common walnut populations.
Abstract
Multiplex polymerase chain reaction (PCR) allows amplification of two or more pair of primers in parallel for amplification of multiple target sequences in a single reaction tube. In this study, we combined existing simple sequence repeat (SSR) markers (nuclear microsatellites) in the novel combination of multiplex PCR to study the population genetics of common walnut from Croatia. From twenty one tested SSR markers, eleven produced satisfactory results in one multiplex PCR. Population genetic results achieved from 15 samples of Croatian common walnut showed moderate genetic variability (average value: He 0.473; Ho 0.568). Our multiplex PCR allowed cost effective work concerning chemicals, plastic ware, device, and working time producing optimal results. The optimized multiplex PCR represented the best combination of eleven SSR primers for genotyping common walnut in a single PCR reaction.
Keywords
SSR markers;
nuclear microsatellites;
Juglans regia L.;
multiplex PCR
-
Ćelepirović,
Division of Genetics, Forest Tree Breeding and Seed Science, Croatian Forest Research Institute, Jastrebarsko, Croatia
E-mail
celepirovic.nevenka@gmail.com

- Karija Vlahović, DNA Laboratory, Department of Forensic Medicine&Criminology, School of Medicine, University of Zagreb, Zagreb, Croatia E-mail monika.karija.vlahovic@mef.hr
- Dounavi, Department of Forest Protection, Forest Research Insitute of Baden-Württemberg, Wonnhaldesttr. 4, 79100 Freiburg, Germany E-mail aikaterini.dounavi@forst.bwl.de
- Ivanković, Division of Genetics, Forest Tree Breeding and Seed Science, Croatian Forest Research Institute, Jastrebarsko, Croatia E-mail mladeni@sumins.hr
Received 11 July 2016 Accepted 18 October 2016 Published 8 November 2016
Views 77563
Available at https://doi.org/10.14214/sf.1674 | Download PDF

1 Introduction
Simple sequence repeats (SSRs), also known as microsatellites, represent DNA regions consisting of two to six nucleotide repeats (Butler 2005a), which are highly polymorphic and uniformly distributed in the eukaryotic genome (Toth et al. 2000). This type of DNA markers is easy to analyze using advanced, cheaper and faster methodologies and automatic capillary electrophoresis. The application across related species increases applicability of SSR markers (Wang et al. 2008). These characteristics make SSRs suitable markers to obtain genetic diversity data of a broad range of organisms (Brinegar 2009). Multiplex PCR is a convenient method for simultaneous amplification of several SSR markers in one reaction. PCR multiplexes can combine previously published SSR markers, which reduce the time and cost of experiment (Holleley 2009). The standardization of multiplex PCR is ensured by uniform reaction conditions, the same amount of template and reagents, and minimized variation produced by human and technical factor (Collins et al. 2004).
Thirty nuclear SSR markers were isolated from black walnut (Juglans nigra L.) library constructed from DNA of ten individuals from USA in 2002 (Woeste et al. 2002). The SSR markers were named with prefix WGA followed by a number and were tested for amplification of the species and cultivars from family Juglandaceae. A considerable number of authors reported their results after applying sets of those SSR markers on different walnut samples and species (Dangle et al. 2005; Victory et al. 2006; Ross-Davis et al. 2008; Ardhya et al. 2010). The same SSR markers were also used for genetic research of national common walnut populations and cultivars in Europe (Foroni et al. 2005; Foroni et al. 2006; Foroni et al. 2007; Cabral et al. 2008; Pollegioni et al. 2011; Ruiz Garcia et al. 2011) and in Asia (Ahmed et al. 2012; Gunn et al. 2010; Ibrahimov et al. 2010; Karimi et al. 2010; Kim et al. 2012; Mahmoodi et al. 2013; Pollegioni et al. 2014; Vahdati et al. 2015; Wang et al. 2008).
Common walnut is an important tree species in Croatia, mainly due to its fruit production. It was planted as individual trees or in small groups in backyards and fruit orchards. Besides planting of the common walnut that grow in Croatia for the centuries, many varieties were imported from neighboring countries, especially after the First World War (Mrva 1995). Although several review articles on PCR multiplex development already exist for forest and agriculture trees (Guichoux et al. 2011), this method is not commonly used for common walnut. Moreover, the genetic variability of Croatian common walnut populations using molecular genetic markers has not been studied so far. The goal of the present study was to set up multiplex PCRs with microsatellite primers to obtain faster and cost effective data for studying the genetic diversity of common walnut populations from Croatia.
2 Materials and methods
2.1 Sample collection and DNA extraction
The samples for DNA extraction were fresh leaves collected from 15 mature common walnut trees of about 25–50 years old. These trees were grown in the municipality of Jastrebarsko (45°40´N, 15°39´E), in Croatia and were planted as urban, ornamental trees. The DNA was extracted according to the protocol of Doyle and Doyle (1987) with the following modifications: hexadecyltrimethylammonium bromide (CTAB) extraction buffer contained 2% polvinylpyrrolidone (PVP). The quality and quantity of isolated DNA was checked on 0.8% agarose gel (Sigma, Germany) using a DNA Ladder (Lambda DNA/HindIII Fragments; Invitrogen, Germany). The DNA concentration was measured fluorometrically (Quantus Fluorimeter, Promega, USA) and ranged from 20.4 to 224.0 ng µl–1.
2.2 PCR conditions, genotyping and genetic analysis
The mostly used SSR markers for genotyping of cultivar and population structure of Juglandaceae were chosen from the literature to study the population structure of common walnut from Croatia. The primer parameters for chosen WGA loci were described in Table 1. SSR loci were amplified in a single PCR reaction with a total volume of 10 µL and in a multiplex PCR reaction with a total volume of 20 µL. The PCR reactions were performed in a PTC-100 Thermal Cycler (MJ Research, Waltham, MA, USA) using fluorescent labelled primers (fluorescent dyes: 6-FAM, VIC, NED, and PET; Applied Biosystems, Life Technologies GmbH). The PCR mixtures contained the following components: 1x PCR buffer, 200 mM each dNTP, 1 U Taq DNA polymerase (TAKARA Co. Ltd, Tokio, Japan), and 1–2 ng template DNA and primers. The primer concentration varied between 0.1 µM and 0.4 µM. The PCR program started with initial denaturation at 95 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, 58 °C for 1 min and 72 °C for 40 s and a final elongation step at 72 °C for 2 min.
| Table 1. Primer sequence information. | |||||||
| Locus name | Primer sequence (5’ to 3’) | Reference | Label | GenBank accession number | Expected size range | Amplified size range | Concentration of primers in multiplex PCR (µM) |
| WGA 001 | F: ATTGGAAGGGAAGGGAAATG R: CGCGCACATACGTAAATCAC | Dangel et al. 2005 | 6 FAM | AY465952 | 180–192 | 186–192 | 0.3 |
| WGA 004 | F: TGTTGCATTGACCCACTTGT R: TAAGCCAACATGGTATGCCA | Dangel et al. 2005 | NED | AY465953 | 228–238 | 228–230 | 0.15 |
| WGA 005 | F: CAGTTTGTCCCACACCTCCT R: AACCCATGGTGAGAGTGAGC | Woeste et al. 2002 | 6 FAM | No data | 240–266 | Multiallele pattern | - |
| WGA 009 | F: CATCAAAGCAAGCAATGGG R: CCATTGCTCTGTGATTGGG | Dangel et al. 2005 | VIC | AY465954 | 229–252 | 236–244 | 0.15 |
| WGA 027 | F: AACCCTACAACGCCTTGATG R: TGCTCAGGCTCCACTTCC | Woeste et al. 2002 | 6 FAM | AY333951 | 192–210 | 202–208 | - |
| WGA 032 | F: CTCGGTAAGCCACACCAATT R: GGACCCAGCTCCTCTTCTCT | Woeste et al. 2002 | VIC | AY333952 | 120–196 | Multiallele pattern | - |
| WGA 071 | F: ACCCGAGAGATTTCTGGGAT R: GGACCCAGCTCCTCTTCTCT | Woeste et al. 2002 | 6 FAM | No data | 136–210 | 205–207 | - |
| WGA 069 | F: TTAGATTGCAAACCCACCCG R: AGATGCACAGACCAACCCTC | Dangel et al. 2005 | PET | AY333953 | 158–180 | 164–184 | 0.15 |
| WGA 089 | F: ACCCATCTTTCACGTGTGTG R: TGCCTAATTAGCAATTTCCA | Dangel et al. 2005 | 6 FAM | AY352440 | 212–220 | 212–220 | 0.25 |
| WGA 118 | F: TGTGCTCTGATCTGCCTCC R: GGGTGGGTGAAAAGTAGCAA | Dangel et al. 2005 | NED | AY479958 | 184–209 | 184–206 | 0.1 |
| WGA 148 | F: GGTGAACTCCCATAGGGGTA R: CCAATGCTACTTGCAGAACC | Woeste et al. 2006 | VIC | DQ307431 | 253–271 | 252–296 | 0.2 |
| WGA 202 | F: CCCCATCTACCGTTGCACTTT R: GCTGGTGGTTCTATCATGGG | Dangel et al. 2005 | VIC | AY479959 | 238–275 | 251–275 | - |
| WGA 204 | F: GGGTCTCGCCTTCTTTTCTT R: CACAGAGAGAAGCACGGGTA | Woeste et al. 2007 | NED | DQ307432 | 172–178 | 174–176 | 0.1 |
| WGA 225 | F: AATCCCTCTCCTGGGCAG R: TGTTCCACTGACCACTTCCA | Dangel et al. 2005 | PET | AY479960 | 190–202 | 192–204 | - |
| WGA 256 | F: TGAAGACAACAAAACTGCGC R: CCGGCATTGTTTCTGAAAAT | Woeste et al. 2009 | PET | DQ307432 | 227–253 | 249–291 | 0.2 |
| WGA 276 | F: CTCACTTTCTCGGCTCTTCC R: GGTCTTATGTGGGCAGTCGT | Dangel et al. 2005 | 6 FAM | AY479961 | 143–192 | 168–194 | - |
| WGA 321 | F: TCCAATCGAAACTCCAAAGG R: GTCCAAAGACGATGATGGA | Dangel et al. 2005 | NED | AY479962 | 222–225 | 223–239 | - |
| WGA 331 | F: TCCCCCTGAAATCTTCTCCT R: CGGTGGTGTAAGGCAAATG | Dangel et al. 2005 | VIC | AY479963 | 273–277 | 271–275 | - |
| WGA 332 | F: ACGTCGTTCTGCACTCCTCT R: GCCACAGGAACGAGTGCT | Foroni et al. 2006 | PET | AY479964 | 214–225 | 215–225 | 0.1 |
| WGA 349 | F: GTGGCGAAAGTTTATTTTTTGC R: ACAAATGCACAGCAGCAAAC | Woeste et al. 2002 | 6 FAM | AY479965 | 258–274 | 262–274 | 0.25 |
| WGA 376 | F: GCCCTCAAAGTGATGAACGT R: TCATCCATATTTACCCCTTTCG | Woeste et al. 2002 | NED | AY479966 | 218–254 | 230–252 | - |
The PCR fragments were separated by capillary electrophoresis on a 3100-Avant Genetic Analyzer (Applied Biosystems), using the GeneScan-500 LIZ (Applied Biosystems) internal size standard. Obtained DNA fragments were sized using GeneMapper software.
Genetic diversity parameters were estimated with GeneAlex software (version 6) (Peakall and Smouse 2006).
3 Results
3.1 Multiplex PCR primer combinations
We tested twenty one previously labeled SSR markers (Table 1) for specificity and sensitivity in single PCRs. The nineteen of them gave positive and repeatable results for all fifteen analyzed samples in the expected size range, except WGA 005 and WGA 032 which produced multilevel pattern (Table 1). The PCR products generated with SSR genotyping produced both homozygote and heterozygote peaks (Fig. 1), which is in accordance to diploid level of common walnut. Our goal was to combine as many SSR markers as possible in one multiplex PCR. Based on single PCR data, we excluded those SSR markers from multiplexing which produce overlapping loci labelled with the same dye (WGA 071, WGA 276, WGA 202, and WGA 027) and excessive stutter bands (WGA 376). The remaining fourteen SSR markers were considered as candidates for the multiplex PCR. Further, we performed the multiplex PCR according to the protocol in the literature (Henegariu et al. 1997) using two different equimolar concentrations of primers: 0.4 µM and 0.15 µM. In contrast to the higher molar concentration (0.4 µM), the lower primer concentrations (0.15 µM) made the peak assignment and the data analysis easier in the multiplex PCR. However, WGA 225, WGA 321, and WGA 331 showed lack of amplification in multiplex PCRs. To achieve even peak heights and peak height balance in multiplex PCR, we empirically optimized the individual primer concentration of eleven SSR markers which varied between 0.1–0.3 µM (Table 1). The remaining eight SSR markers which were excluded from the multiplex PCR were possible to be combined into two smaller multiplex PCRs: one five-primer-set (WGA 027, WGA 225, WGA 276, WGA 331, and WGA 321) and one three-primer-set (WGA 071, WGA 202, and WGA 376) (data not shown).
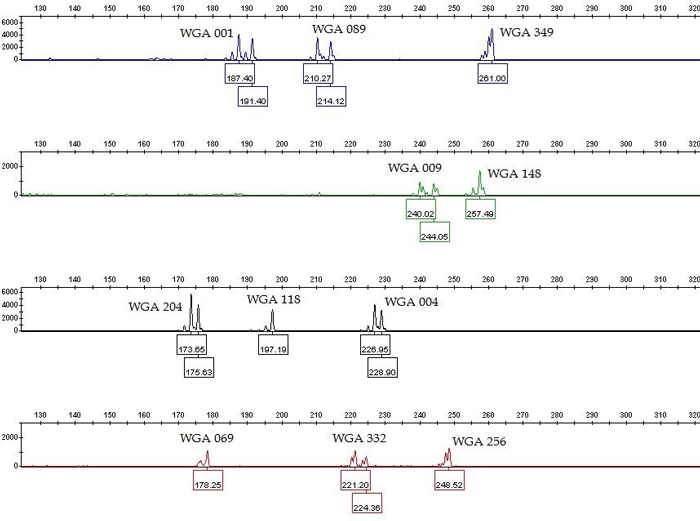
Fig. 1. Genotype profile (electropherogram) of the eleven microsatellite loci (SSR markers) of common walnut generated by multiplex PCR. The x-axis represents DNA fragment size in base pairs, and the y-axis represents fluorescence units. The SSR profile is displayed in the blue (6-FAM labeled primers), green (VIC labeled primers), yellow (NED labeled primers) and red (PET labeled primers) channels of a five-color fluorescent system, with the orange (LIZ labeled fragments) channel being used for a size marker (not shown). View larger in new window/tab.
3.2 Genotyping data
A summary of gene diversity parameters of 15 common walnut samples at each of eleven analyzed loci is presented in Table 2. Genetic diversity was moderate for most of the investigated loci. In total, 42 alleles were found. All markers were polymorphic with a maximum number of alleles Na = 7 (WGA 069) and a minimum Na = 2 (WGA 004 and WGA 204) with an average value of 3.818. Markers WGA 001, WGA 069 and WGA 148 showed excess of homozygotes. Observed heterozygosity ranged from 0.067 to 0.667 with an average of 0.473. Fixation index (FIS) indicated excess of heterozygosity in five loci (ranged from –0.034 to –0.132) and a deficiency of heterozygotes in six loci (ranged from 0.016 to 0.668). Only three of them indicated significant departure from Hardy-Weinberg equilibrium.
| Table 2. Gene diversity parameters of eleven common walnut SSR markers, sample size (N), number of observed alleles (Na), effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He), fixation index (FIS), and Hardy-Weinberg equilibrium (HW). | |||||||
| Locus | N | Na | Ne | Ho | He | FIS | HW |
| WGA 001 | 15 | 4 | 2,514 | 0,200 | 0,602 | 0,668 | *** |
| WGA 004 | 15 | 2 | 1,923 | 0,533 | 0,480 | –0,111 | ns |
| WGA 009 | 15 | 3 | 2,761 | 0,533 | 0,638 | 0,164 | ns |
| WGA 069 | 15 | 7 | 4,091 | 0,333 | 0,756 | 0,559 | * |
| WGA 089 | 15 | 4 | 3,103 | 0,667 | 0,678 | 0,016 | ns |
| WGA 118 | 15 | 4 | 2,571 | 0,667 | 0,611 | –0,091 | ns |
| WGA 148 | 15 | 6 | 2,586 | 0,400 | 0,613 | 0,348 | *** |
| WGA 204 | 15 | 2 | 1,069 | 0,067 | 0,064 | –0,034 | ns |
| WGA 256 | 15 | 4 | 2,432 | 0,667 | 0,589 | –0,132 | ns |
| WGA 332 | 15 | 3 | 2,761 | 0,667 | 0,638 | –0,045 | ns |
| WGA 349 | 15 | 3 | 2,381 | 0,467 | 0,580 | 0,195 | ns |
| Mean | 15 | 3,818 | 2,563 | 0,473 | 0,568 | 0,140 | |
| SE | 0 | 0,464 | 0,222 | 0,062 | 0,054 | 0,084 | |
| ns = not significant, * P<0.05, ** P<0.01, *** P<0.001 | |||||||
4 Discussion
Multiplex PCR enables quick and cost effective amplification of samples with more than one primer pair in one reaction (Edward and Gibbs 1994). Theoretically, the number of primer pairs in single multiplex mix is limited by the number and range of fragment size (Culley et al. 2013). In our work, twenty one SSR markers previously used to study the genetic diversity of populations and cultivars of common walnut (Dangel et al. 2005; Ross-Davis et al. 2008; Woeste et al. 2002) were used for genotyping of 15 samples of common walnut from Croatia. We showed that the highest number of SSR markers combinable in one multiplex PCR was eleven primer pairs. The remaining eight SSR markers were possible to be combined in two multiplex PCRs, with five and three SSR markers, respectively. Also, three SSR markers did not amplify in multiplex PCR, probably because of competition between primers (Butler 2005b; Henegariu et al. 1997). The limiting factor for choosing the candidate primer pairs for multiplex PCR was labelling of primers with the available fluorescent dyes for genotyping with ABI PRISM® 3100-Avant Genetic Analyzer. Particularly, for multiplex formation, we gave special attention to avoid overlapping of size ranges of the fragments labelled with the same fluorescent dye. With the optimization of primer concentrations, we achieved peak height balance at heterozygous SSR loci. The developed multiplex panel could be enlarged by modifying of the cycling conditions, the thoughtfully fluorescent labelling of SSR markers and broadening of the tested size range.
Information of genetic diversity and population structure are important to understand long term variability and adaptability of populations (Rajora et al. 2001). The study of molecular diversity of common walnut is essential in the preservation of its genetic resources. The common walnut is species that predominantly growth in human settlement, so its selection and distribution is directly affected by humans. Traditionally managed common walnut populations may serve as reservoir for genetic variation (Gun et al. 2010). Analyses of fifteen samples of Croatian walnut population from small geographic area showed moderate polymorphic level and genetic diversity. The observed average number of alleles per locus is highly dependent on sample size. Accordingly, the average number of alleles of Croatian samples was lower compared with those in literature in which more individual trees were studied but the average number of effective alleles was consistent with the published data (Pollegioni et al. 2009; Pollegioni et al. 2011; Ruiz-Garcia et al. 2011). The investigation of Croatian walnut population, which is small but complex population considering the mixing of different genotypes over the centuries, will provide insight into effects of traditional management on its population genetics.
Acknowledgments
Funding was provided by Ministry of Science, Education and Sports of the Republic of Croatia. to research project Tree breeding and forest seed science (024-0242108-2099) and by Croatian forest L.t.d. to research project Genetic variability of Families of Common Walnut.
References
Ahmed N., Mir J.I., Mir R.R., Rather N.A., Rashid R., Wani S.H., Shafi W., Mir H., Sheikh M.A. (2012). SSR and RAPD analysis of genetic diversity in walnut (Juglans regia L.) genotypes from Jammu and Kashmir, India. Physiology and Molecular Biology of Plants 18(2): 149–160. http://dx.doi.org/10.1007/s12298-012-0104-z.
Aradhya M., Woeste K., Velasco D. (2010). Genetic diversity, structure and differentiation in cultivated walnut (Juglans regia L.). Acta Horticulturae 861: 127–132. http://dx.doi.org/10.17660/ActaHortic.2010.861.16.
Brinegar C. (2009). Assessing evolution and biodiversity in plants at the molecular level. KUSET 5(2): 149–159.
Butler J.M. (2005a). Forensic DNA typing: biology, technology, and genetics of STR markers. Elsevier Academic Press, Burlington, MA.
Butler J.M. (2005b). Constructing STR multiplex assays. Methods in Molecular Biology 297: 53–65.
Cabral E.M.F.D.M. (2008). Caracterização Molecular de Variedades de Nogueira (Juglans regia L.) Portuguesas. [Molecular characterization of Portuguese walnut varieties (Juglans regia L.)]. Dissertação, Faculdade de Farmácia Universidade do Porto, Porto. [Dissertation, Faculty of Pharmacy University of Porto, Porto]. [In Portuguese].
Collins P.J., Hennessy L.K., Leibelt C.S., Roby R.K., Reeder D.J., Foxall P.A. (2004). Developmental validation of a single-tube amplification of the 13 CODIS STR loci, D2S1338. D19S433. and amelogenin: the AmpFlSTR® Identifiler® PCR Amplification Kit. Journal of Forensic Sciences 49(6): 1265–1277. http://dx.doi.org/10.1520/JFS2002195.
Culley T.M., Stamper T.I., Stokes R.L., Brzyski J.R., Hardiman N.A., Klooster M.R., Merritt B.J. (2013). An efficient technique for primer development and application that integrates fluorescent labeling and multiplex PCR. Applications in Plant Sciences 1(10): 1300027. http://dx.doi.org/10.3732/apps.1300027.
Dangle G.S., Woeste K., Aradhya M.K., Koehmstedt A., Simon C., Potter D., Leslie C.A., McGranahan G. (2005). Characterization of 14 SSR markers for genetic analysis and cultivar identification of walnut. Journal of the American Society for Horticultural Science 130(3): 348–354.
Doyle J.J., Doyle J.L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15.
Edwards M.C., Gibbs R.A. (1994). Multiplex PCR: advantages, development, and applications. Genome Research 3(4): 65–75. http://genome.cshlp.org/content/3/4/S65.long.
Foroni I., Rao R., Woeste K., Galliteli M. (2005). Characterisation of Juglans regia L. with SSR markers and evaluation of genetic relationships among cultivars and the’Sorrento’landrace. The Journal of Horticultural Science and Biotechnology 80(1): 49–53. http://dx.doi.org/10.1080/14620316.2005.11511890.
Foroni I., Rao R., Woeste K. (2006). Molecular characterization of Juglans regia L. cultivars with SSR markers. Acta Horticulturae 705: 207-213. http://dx.doi.org/10.17660/ActaHortic.2005.705.25.
Foroni I., Woeste K., Monti L.M., Rao R. (2007). Identification of ‘Sorrento’walnut using simple sequence repeats (SSRs). Genetic Resources and Crop Evolution 54(5): 1081–1094. http://dx.doi.org/10.1007/s10722-006-9187-0.
Guichoux E., Lagache L., Wagner S., Chaumeil P., Léger P., Lepais O., Lepoittevin C., Malausa T. (2011) Current trends in microsatellite genotyping. Molecular Ecology Resources 11(4): 591–611. http://dx.doi.org/10.1111/j.1755-0998.2011.03014.x.
Gunn B.F., Aradhya M., Salick J.M., Miller A.J., Yongping Y., Lin L., Xian H. (2010). Genetic variation in walnuts (Juglans regia and J. sigillata; Juglandaceae): species distinctions, human impacts, and the conservation of agrobiodiversity in Yunnan, China. American Journal of Botany 97(4): 660–671. http://dx.doi.org/10.3732/ajb.0900114.
Henegariu O., Heerema N.A., Dlouhy S.R., Vance G.H., Vogt P.H. (1997). Multiplex PCR: critical parameters and step-by-step protocol. BioTechniques 23(3): 504–511.
Holleley C.E., Geerts P.G. (2009). Multiplex Manager 1.0: a cross-platform computer program that plans and optimizes multiplex PCR. BioTechniques 46(7): 511–517. http://dx.doi.org/10.2144/000113156.
Ibrahimov Z., McGranahan G.H., Leslie C.A., Aradhya M. (2010). Genetic diversity in walnut (Juglans regia) from the caucasus nation of azerbaijan. Acta Horticulturae 861: 163–170. http://dx.doi.org/10.17660/ActaHortic.2010.861.21.
Karimi R., Ershadi A., Vahdati K., Woeste K. (2010). Molecular characterization of Persian walnut populations in Iran with SSR markers. HortScience 45(9): 1403–1406.
Kim H.B., Jeon J.H., Han A.R., Lee Y., Jun S.S., Kim T.H., Cho G.H., Park P.B. (2012). Genetic evaluation of domestic walnut cultivars trading on Korean tree markets using SSR markers. African Journal of Biotechnology 11(29): 7366–7374.
Mahmoodi R., Rahmani F., Rezaee R. (2013). Genetic diversity among Juglans regia L. genotypes assessed by morphological traits and SSR markers. Spanish journal of agricultural research 11(2): 431–437. http://dx.doi.org/10.5424/sjar/2013112-3445.
Mrva F. (1984). Mogućnost proširenja uzgoja domaćeg oraha. [The possibilitiey of extending cultivation of local walnuts]. Glasnik 5: 621–633.
Peakall R., Smouse P.E. (2006). GenAlex 6: genetic analysis in excel. Population genetic software for teaching and research. Molecular Ecology Resources 6: 288–295. http://dx.doi.org/10.1111/j.1471-8286.2005.01155.x.
Pollegioni P., Woeste K., Major A., Scarascia Mugnozza G., Malvolti M.E. (2009). Characterization of Juglans nigra (L.), Juglans regia (L.) and Juglans × intermedia (Carr.) by SSR markers: a case study in Italy. Silvae Genetica 58(1–2): 68–78.
Pollegioni P., Woeste K., Olimpieri I., Marandola D., Cannata F., Malvolti M.E. (2011). Long-term human impacts on genetic structure of Italian walnut inferred by SSR markers. Tree Genetics & Genomes 7(4): 707–723. http://dx.doi.org/10.1007/s11295-011-0368-4.
Pollegioni P., Woeste K.E., Chiocchini F., Olimpieri I., Tortolano V., Clark J., Hemery G.E., Mapelli S., Malvolti M.E. (2014). Landscape genetics of Persian walnut (Juglans regia L.) across its Asian range. Tree Genetics & Genomes 10(4): 1027–1043. http://dx.doi.org/ 10.1007/s11295-014-0740-2.
Rajora O.P., Mosseler A. (2001). Challenges and opportunities for conservation of forest genetic resources. Euphytica 118(2): 197–212. http://dx.doi.org/10.1023/A:1004150525384.
Ross-Davis A., Woeste K.E. (2008). Microsatellite markers for Juglans cinerea L. and their utility in other Juglandaceae species. Conservation Genetics 9(2): 465–469. http://dx.doi.org/10.1007/s10592-007-9337-8.
Ruiz-Garcia L., Lopez-Ortega G., Fuentes Denia A., Frutos Tomas D. (2011). Identification of a walnut (Juglans regia L.) germplasm collection and evaluation of their genetic variability by SSR markers. Spanish Journal of Agricultural Research 9(1): 179–192. http://dx.doi.org/10.5424/sjar/20110901-227-10.
Tóth G., Gáspári Z., Jurka J. (2000). Microsatellites in different eukaryotic genomes: survey and analysis. Genome Research 10(7): 967–981. http://dx.doi.org/10.1101/gr.10.7.967.
Vahdati K., Pourtaklu S.M., Karimi R., Barzehkar R., Amiri R., Mozaffari M., Woeste K. (2015). Genetic diversity and gene flow of some Persian walnut populations in southeast of Iran revealed by SSR markers. Plant Systematics and Evolution 301(2): 691–699. http://dx.doi.org/10.1007/s00606-014-1107-8.
Victory E.R., Glaubitz J.C., Rhodes Jr O.E., Woeste K.E. (2006). Genetic homogeneity in Juglans nigra (Juglandaceae) at nuclear microsatellites. American Journal of Botany 93(1): 118–126. http://dx.doi.org/10.3732/ajb.93.1.118.
Wang H., Pei D., Gu R.S., Wang B.Q. (2008). Genetic diversity and structure of walnut populations in central and southwestern China revealed by SSR markers. Journal of the American Society for Horticultural Science 133(2): 197–203.
Woeste K., Burns R., Rhodes O., Michler C. (2002). Thirty polymorphic nuclear microsatellite loci from black walnut. Journal of Heredity 93(1): 58–60. http://dx.doi.org/10.1093/jhered/93.1.58.
Total of 35 references.

