Homogenous genetic structure in populations of Taxus baccata with varied proportions of male and female individuals
Litkowiec M., Plitta-Michalak B. P., Lewandowski A., Iszkuło G. (2015). Homogenous genetic structure in populations of Taxus baccata with varied proportions of male and female individuals. Silva Fennica vol. 49 no. 4 article id 1236. https://doi.org/10.14214/sf.1236
Highlights
- Polish populations of Taxus baccata showed a high level of genetic diversity within populations and moderate genetic differentiation between them after nSSR marker testing
- No significant differences in the genetic variation between T. baccata male and female individuals were observed, and microsatellite loci neutrality was verified
- Determining the sex ratio in T. baccata populations is not essential to develop a clear understanding of genetic differentiation and diversity within and between populations of this species.
Abstract
English yew (Taxus baccata L.) is a strictly outcrossing and dioecious species whose populations are small and isolated. It is known that sex ratios may vary in natural populations due to local environmental conditions or stochastic events. However, unbalanced sex ratios may have negative impacts on genetic diversity through enhanced genetic drift and inbreeding. The present study represents one of the first attempts to compare the genetic variation at microsatellite loci within and between populations with different gender proportions. Our results indicated that there were no significant correlations between sex ratio and the extent of genetic variation in different populations. All populations exhibited high levels of genetic diversity. Additionally, the genetic structure was characterized separately in male and female individuals. Statistical analyses of the set estimators describing the genetic structure of male and female individuals of T. baccata revealed no significant differences between the two groups. Molecular analysis verified that microsatellite nuclear loci neutrality developed for T. baccata, as there were no significant differences in the genetic variation between males and females and no evidence for any outlier loci using coalescent and hierarchical Bayesian simulations. The results demonstrate that ignoring biased sex ratios in T. baccata populations had no effect on the assessment of genetic differentiation and genetic diversity within and between populations of this species. These results are discussed with regards to the practical application of molecular markers in conservation programs.
Keywords
genetic diversity;
dioecy;
English yew;
Poland;
sex ratio;
nSSR markers
-
Litkowiec,
Institute of Dendrology, Polish Academy of Sciences, Parkowa 5, 62-035 Kórnik, Poland
E-mail
mlit@man.poznan.pl

- Plitta-Michalak, Institute of Dendrology, Polish Academy of Sciences, Parkowa 5, 62-035 Kórnik, Poland E-mail beata-plitta@wp.pl
- Lewandowski, Institute of Dendrology, Polish Academy of Sciences, Parkowa 5, 62-035 Kórnik, Poland E-mail alew@man.poznan.pl
- Iszkuło, Institute of Dendrology, Polish Academy of Sciences, Parkowa 5, 62-035 Kórnik, Poland & University of Zielona Góra, Faculty of Biological Sciences, Prof. Z. Szafrana 1, 65-516 Zielona Góra, Poland E-mail iszkulo@man.poznan.pl
Received 18 August 2014 Accepted 15 June 2015 Published 6 July 2015
Views 172757
Available at https://doi.org/10.14214/sf.1236 | Download PDF

Supplementary Files
1 Introduction
Dioecy is a relatively rare mating system in plants characterized by individual plants being distinctly male or female. Dioecious species comprise approximately 10% of all known species of land plants, 4–6% of which are angiosperm species (Givnish 1980; Renner and Ricklefs 1995; Soza et al. 2012; Barrett and Hough 2013). Phylogenetic studies of angiosperm dioecious plants indicate a low speciation level and higher extinction risk compared to cosexual species (Heilbuth 2000; Vamosi and Vamosi 2005). The increased extinction risk is often linked to an insufficient number of individuals of the opposite sex, particularly in small and isolated populations (Pannell and Barrett 1998). However, dioecy can be beneficial from an evolutionary point of view due to the genetic advantages associated with guaranteed outbreeding (Freeman et al. 1997).
The sex ratio parameter is frequently applied to describe the population structure of dioecious species. The theoretical sex ratio should be 1:1 (Fisher 1930) as the result of matings between individuals of the opposite sex. Among dioecious plants, there are frequent significant equality deviations in natural populations (see review Sinclair et al. 2012), which could negatively affect their genetic diversity. Environmental factors (Mercer and Eppley 2010) and stochastic events, particularly in small isolated populations (Engen et al. 2003), lead to variations in gender proportions among populations. In populations with unbalanced sex ratios, the effective population size is low, possibly leading to decreased allelic diversity owing to enhanced genetic drift (Hedrick 2000). Moreover, in such dioecious species populations, diminished reproductive efficiency due to an insufficient number of individuals of the opposite sex also affects genetic diversity (Eppley 2005; Vandepitte et al. 2009).
English yew (Taxus baccata L.) is a canonical example of a strictly outcrossing and dioecious species, though the opposite sexes have the same or nearly the same characteristics (Svenning and Magård 1999; Thomas and Polwart 2003; Iszkuło et al. 2009). Female T. baccata individuals grow slower than males, which are typically taller and have a larger trunk diameter than females (Iszkuło et al. 2009; Cedro and Iszkuło 2011). Iszkuło et al. (2009) reported that the percentage of females increases with higher precipitation, and sex ratios only reach a balance in populations located in areas with high annual rainfall. Collectively, these factors contribute to changes in sex structure (i.e., decreased number of females) observed in ageing populations (Iszkuło et al. 2009). Several studies have calculated sex ratios in natural T. baccata populations and found that they vary from 40% female (see review Thomas and Polwart 2003) to 71% female (Hilfiker et al. 2004a). Most studies on the genetic structure of small and fragmented T. baccata populations in Europe utilized different system markers and demonstrated a high level of genetic variation and significant genetic differentiation between populations, the latter resulting from genetic drift and inbreeding (Lewandowski et al. 1995; Cao et al. 2004; Hilfiker et al. 2004b; Dubreuil et al. 2010; Myking et al. 2009; Trӧber and Ballian 2011; Chybicki et al. 2011; 2012; Litkowiec et al. 2013). However, those studies were conducted without consideration of the gender proportions in analysed populations.
Due to increased inbreeding and genetic drift, both of which have negative genetic impacts on populations, it is important to investigate and compare genetic variation within and between a large number of populations with unbalanced sex ratios. For this investigation of T. baccata populations, five highly polymorphic nuclear microsatellite (nSSR) markers were selected. Specifically, our aims were to (1) describe the values of parameters used to characterize genetic structure in male and female individuals, (2) determine whether microsatellite markers have a neutral character, and (3) determine the correlation between sex ratios and genetic variation in populations.
2 Material and methods
2.1 Plant material
Needles were collected from 26 natural T. baccata populations in Poland (Table 1). These populations covered the entire area of Poland and were part of a conservation and restoration program launched in 2006 (Fig. 1). The sex ratios (% female) in the studied populations were based on data collected by Kostrzyca Forest Gene Bank. The presence of micro- and macro-strobili and seeds or seed envelope remains were used to determine the sex of each individual. Groups of male and female individuals were defined in each population based on this information. In total, 2198 individuals, including 1040 males and 1158 females, were subjected to microsatellite analysis.
| Table 1. The location, code, and percentage of females in the studied populations and the number of male and female Taxus baccata individuals analysed. | |||||
| Population | Pop. Number | Longitude/Latitude | % females a | Analyzed males | Analyzed females |
| Leszno | 1 | 53°46´N/20°52´E | 45 | 50 | 50 |
| Choczewskie Cisy | 2 | 54°46´N/17°45´E | 47 | 36 | 46 |
| Cisy nad Czerską Strugą | 3 | 53°45´N/17°57´E | 46 | 40 | 47 |
| Jelenia Góra | 4 | 53°35´N/18°06´E | 50 | 38 | 36 |
| Wierzchlas | 5 | 53°30´N/18°06´E | 33 | 50 | 50 |
| Cisy w Czarnem | 6 | 53°44´N/16°58´E | 49 | 48 | 48 |
| Cisy Rokickie | 7 | 53°45´N/14°51´E | 64 | 49 | 50 |
| Nadleśnictwo Rokita | 8 | 53°42´N/14°51´E | 63 | 25 | 46 |
| Boleszkowice | 9 | 52°42´N/14°33´E | 55 | 37 | 55 |
| Bogdaniec | 10 | 52°41´N/15°04´E | 32 | 32 | 32 |
| Książ | 11 | 50°50´N/16°16´E | 37 | 43 | 37 |
| Cisowa Góra | 12 | 50°32´N/16°42´E | 53 | 50 | 50 |
| Cisy koło Barda | 13 | 50°32´N/16°39´E | 51 | 50 | 50 |
| Nowy Waliszów | 14 | 50°18´N/16°45´E | 51 | 50 | 50 |
| Cisy nad Liswartą | 15 | 50°44´N/18°46´E | 54 | 34 | 55 |
| Cisy w Hucie Starej | 16 | 50°33´N/19°12´E | 48 | 26 | 27 |
| Jasień | 17 | 50°59´N/19°33´E | 55 | 41 | 52 |
| Radomice | 18 | 50°44´N/20°39´E | 68 | 29 | 65 |
| Mogilno | 19 | 49°39´N/20°49´E | 58 | 38 | 53 |
| Wadernik | 20 | 49°29´N/21°38´E | 51 | 26 | 30 |
| Igiełki | 21 | 49°30´N/21°38´E | 54 | 30 | 39 |
| Kretówki | 22 | 49°44´N/21°53´E | 40 | 58 | 39 |
| Malinówka | 23 | 49°41´N/21°55´E | 44 | 50 | 50 |
| Pasmo Łysej Góry | 24 | 49°33´N/21°34´E | 55 | 42 | 41 |
| Serednica | 25 | 49°28´N/22°31´E | 46 | 31 | 26 |
| Cisy na Górze Jawor | 26 | 49°17´N/22°17´E | 55 | 38 | 34 |
| a Based on data collected by Kostrzyca Forest Gene Bank | |||||
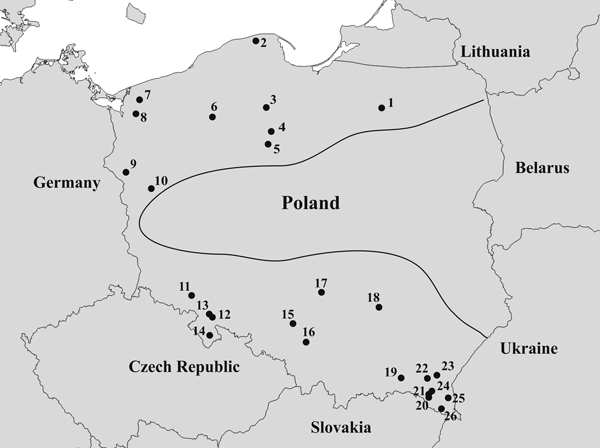
Fig. 1. Map of the geographical locations of the 26 Taxus baccata populations included in this study. Codes representing the named populations are listed in Table 1, and the borders of the natural T. baccata distribution in Poland are indicated with a black line.
2.2 DNA extraction and microsatellite genotyping
Genomic DNA was extracted from needles using a modified CTAB protocol described by Dumolin et al. (1995). The identification of suitable markers was based on their ability to provide repeatable, high quality results, sufficient polymorphism, and unambiguous allele binding of eight nuclear microsatellite markers originally identified in T. baccata by Dubreuil et al. (2008). The final set of loci included five nSSRs (tax23, tax26, tax31, tax36, tax92).
PCR reactions were performed in one multiplex reaction and two single reactions. Three nSSR loci (tax26, tax36 and tax92) were simultaneously amplified in a multiplex reaction using a Qiagen Multiplex PCR kit (Qiagen, Hilden, Germany). PCR multiplex reactions were performed in a total volume of 10 µl composed of 5 µl Qiagen Multiplex Master Mix (2X), 0.2 µl primer mix (20 µM), 1 µl Q-Solution (5X), 0.8 µl RNase-Free water, and 3 µl DNA template (approximately 10–20 ng). PCR reactions for amplifying loci tax23 and tax31 were performed in a total volume of 10 µl, containing 10–20 ng DNA template, 1X PCR, pH 8.3 (Novazym, Poland), 25 mM MgCl2, 0.2 mM dNTPs, 0.8 µM of each primer, 10 ng/µl BSA, and 1.25 U VivaTaq polymerase (Novazym, Poznan, Poland). PCR conditions for both the single and multiplex reactions were as follows: an initial denaturation step at 95 °C for 15 min, followed by 10 touchdown cycles at 94 °C for 30 s, 30 s at 60 °C (–1 °C/ cycle), 40 s at 72 °C, 30 cycles at 94 °C for 30 s, 50 s at 50 °C, 40 s at 72 °C, and a final extension at 72 °C for 7 min. The fluorescently labelled PCR products and a size standard (GeneScan 500 LIZ) were separated on a capillary sequencer ABI 3130 (Life Technologies, Carlsbad, CA, USA). The alleles were identified based on their size using GeneMapper software (ver. 4.0; Life Technologies).
2.3 Data analysis
Genotypic linkage disequilibrium for all microsatellite loci was verified in each population and across all populations based on a permutation test using Arlequin software (Excoffier et al. 2005). The genetic diversity within and among populations was estimated based on the following parameters: total number of alleles (A); allelic richness (AR25) corrected for a minimum sample size of 25 individuals; observed heterozygosity (Ho); and unbiased expected heterozygosity (He), all of which were computed using FSTAT v 2.9.3 (Goudet 2001). Null alleles are known to be present in the nSSR markers used in this study (Dubreuil et al. 2008). Therefore, the frequencies of null alleles (N0) in each population were estimated based on the Individual Inbreeding Model (IIM) with a Gibbs sampler 105 iterations using INEST 1.0 software (Chybicki and Burczyk 2009). All parameters describing genetic variation in a population were separately estimated for male and female individuals.
The global genetic differentiation (Fst) among overall samples was assessed as Fst values (Weir and Cockerham 1984) with FSTAT v. 2.9.3 (Goudet 2001). Taking into account the presence of null alleles at all loci, FreeNA software was used to estimate Fst using the ENA (Excluding Null Alleles, FstENA) correction method (Chapuis and Estoup 2007). The bootstrap 95% confidence intervals (CI) for global Fst values were calculated using 20 000 replicates over loci.
FSTAT was used to compare grouped male and female samples based on various genetic variation parameters. Statistically significant differences between the overall means of the standard parameters describing genetic structure were determined using a permutation test (10 000 permutations) within FSTAT. Several parameters, including AR, A, Fst, mean of He and Ho, were compared. Due to software limitations, not all genetic diversity parameters were statistically analysed in FSTAT. Analysis of variance (ANOVA) was used to determine significant differences (p-value ≤ 0.05) between the means of male and female frequencies of null alleles.
The genetic relationship among populations was further analysed by principle coordinate analysis (PCoA) using GenAlEx v. 6 (Peakall and Smouse 2006) based on FstENA values generated by FreeNA with an ENA correction.
Outlier loci, i.e., markers potentially under selection, were detected using two different approaches to minimize false positive detection. First, the Beaumont and Nichols method (1996) in the Lositan Selection Workbench (Antao et al. 2008) was used to determine whether microsatellite loci were outliers. The method uses a coalescent simulation approach based on an island model for migration to identify outlier loci displaying unusually high (i.e., putatively under directional selection) and low (i.e., potentially under stabilizing selection) values of Fst by comparing observed Fst values with values expected under neutrality. Loci with a high Fst value are putatively under directional selection, while loci with a low Fst value are considered to potentially be under stabilizing selection. The evaluation was performed using 50 000 iterations, stepwise mutation models, and all loci. The mean neutral Fst was used as a preliminary value. Loci lying outside a 99.5% confidence interval were considered to be potentially under selection rather than neutral. Second, the hierarchical Bayesian simulation (Beaumont and Balding 2004) in Bayescan 2.1 software (Foll and Gaggiotti 2008) was used to detect non-neutral loci. This method is based on the decomposition of locus-population Fst coefficients into a population-specific component (beta), shared by all loci and a locus-specific component (alpha) shared by all populations using a logistic regression model. Analyses were performed using a default set of parameters. To avoid false positives, the higher prior odds (1000) were used. The scale based on Bayes factor described by Jeffreys (1961) was used to consider a locus under selection.
Regression analyses were used to infer relationships between sex ratios and genetic diversity (A, AR25, He, Ho). Analyses were conducted using JMP® statistical software (SAS Institute Inc., Cary, NC, USA, 1989–2007).
3 Results
All nSSR loci were polymorphic, and there was no evidence detected to indicate linkage disequilibrium. The examined populations and grouped individuals of male and female T. baccata samples exhibited a high level of genetic diversity, and the level was similar within populations and for each sex. The genetic diversity parameters for whole populations and the separate male and female samples are displayed in Table 2. The mean number of alleles per population (A) ranged from 2.2 in the Bogdaniec (10) to 14.6 in the Cisy koło Barda (13), with an overall average of 9.4 among all populations. The mean number of alleles ranged from 2.2 in male samples from the Bogdaniec (10) to 14.0 in male samples from the Cisowa Góra (12) and Cisy koło Barda (13), with an overall average of 9.4 among all males. A similar trend was observed in the female samples, with the lowest number of alleles in females from the Bogdaniec (10; A = 2.2) and the highest in females from Cisy koło Barda (13; A = 15.2), with an overall average of 9.5 in all females. Allelic richness (AR) was estimated for the smallest number of individuals (25) with an assigned sex, and the overall value was 7.8; the highest value was in Cisowa Góra (12; AR = 10.9) and Cisy koło Barda (13; AR = 11.0), while the lowest value was in the Bogdaniec (10; AR = 2.2). The average AR for male samples was 7.7. The lowest AR was observed in Bogdaniec (10; AR = 2.2), while the highest values, 11.1 and 11.4, were in the Cisowa Góra (12) and Cisy koło Barda (13) populations, respectively. The values of these parameters in female groups ranged from 2.2 in the Bogdaniec (10) to 10.9 in the Książ (11).
| Table 2. Parameters used to estimate the genetic structure of the studied Taxus baccata populations averaged across five nSSR loci. A – average number of alleles, AR25 –allelic richness, He – expected heterozygosity, Ho – observed heterozygosity, N0 – null allele frequency. View in new window/tab. |
The level of overall observed heterozygosity (Ho) and per population (mean 0.483, range: 0.263–0.591) was considerably lower than the level of expected heterozygosity (He) (mean 0.731, range: 0.441- 0.839). The average He value in the male and female grouped samples was similar at 0.731 and 0.730, respectively. He in males ranged from 0.441 in the Bogdaniec (10) to 0.844 in the Książ (11) and from 0.441 in the Bogdaniec (10) to 0.845 in the Cisy koło Barda (13) in the female groups. The average Ho value in male and female grouped samples was much lower than the He averages (Ho: 0.487 vs. He: 0.479). The lowest Ho level for both male and female samples (Ho = 0.263) was in the Bogdaniec (10) population, while the highest Ho values for males was in the Książ (11) (Ho = 0.614) and in the Cisy w Czarnem (6) (Ho = 0.590) for females.
Null alleles (N0) were detected at moderate frequencies per population, ranging from 8.8% in the Nowy Waliszów (14) to 18.9% in the Jasień (17), with an overall mean of 14.2%. Similar N0 frequencies were obtained in most of the male and female samples (average values: 13.8% vs. 14.7%). In some cases, however, the null allele frequencies for male and female individuals within a single population significantly varied. For example, the N0 frequencies were 5.5% and 12.1% for males and females, respectively, in the Nowy Waliszów (14) population (Table 2).
Given the presence of N0, the global genetic differentiation (Fst) level was estimated separately in males and females with and without ENA correction. These two approaches for estimating the Fst level gave similar results. Fst values not using ENA vs. Fst using ENA for males were 0.152 [confidence interval, CI = 0.134–0.164] and 0.134 [CI = 0.123–0.146], respectively, and were 0.149 [CI = 0.134–0.170] and 0.135 [CI = 0.121–0.151], respectively, for females. These results indicate that the classical measures of population differentiation are slightly biased by the presence of null alleles.
No significant difference (p-values > 0.1) were observed in the averages of the genetic variation estimators between male and female individuals (Table 3). The difference in the Fst in male and female samples (0.152 and 0.149, respectively) was not statistically significant (p-value = 0.901) (Table 3). These relationships are well illustrated in the PCoA plot where male and female individuals share a common subgroup for each population. The first and second principal coordinates accounted for 26.51% and 19.51% of the total variation, respectively (Fig. 2).
| Table 3. Parameters used to estimate the level of genetic diversity and differentiation in male and female individuals A – average number of alleles, AR25 – allelic richness, He – expected heterozygosity, Ho – observed heterozygosity, N0 – null allele frequency, Fst –genetic differentiation, p–values of the statistical significance of differences between the two sample groups. | |||
| Parameter | Female samples | Male samples | p-value |
| A | 9.512 | 9.469 | 0.945 |
| AR25 | 7.769 | 7.744 | 0.965 |
| He | 0.730 | 0.731 | 0.911 |
| Ho | 0.479 | 0.487 | 0.876 |
| N0 | 0.146 | 0.137 | 0.275 |
| Fst | 0.149 | 0.152 | 0.901 |
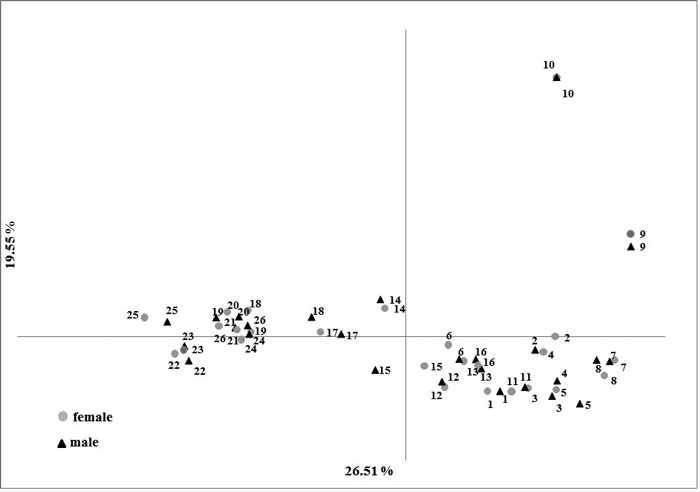
Fig. 2. Results of Principal Component Analysis based on five nuclear SSR markers in male and female Taxus baccata individuals.
The outlier tests performed using both analytical approaches (Lositan, Bayescan) showed no evidence for any outlier loci among the five microsatellite loci studied. This analysis confirmed that the loci have a presumed neutral character (Fig. S1, Table S1, available as a supplementary file).
Regression analyses between sex ratio and the amount of genetic diversity (A, AR25, He, Ho) in the T. baccata populations were estimated, and there were no significant correlations between sex ratio and mean number of alleles per population (r2 = 0.024, p = 0.445). Furthermore, no significant correlations were found between sex ratio and allelic richness (r2 = 0.010, p = 0.611), sex ratio and expected heterozygosity (r2 = 0.034, p = 0.364), and sex ratio and observed heterozygosity (r2 = 0.071, p = 0.188).
4 Discussion
This study represents one of the first attempts to compare the level of genetic variation and genetic differentiation in T. baccata populations with unbalanced sex ratios using nuclear microsatellite analysis. Moreover, the values for a set of parameters characterizing the genetic structure were estimated for individuals of each sex. This type of comparison is valuable because there are known biased sex ratios in T. baccata, which may have a negative genetic impact on the small and fragmented populations.
Several earlier studies have reported a high level of genetic variation and a moderate level of genetic differentiation in natural T. baccata populations (Lewandowski et al. 1995; Hilfiker et al. 2004b; Myking et al. 2009; Zarek 2009; Dubreuil et al. 2010; González-Martínez et al. 2010; Trӧber and Ballian 2011; Chybicki et al. 2011, 2012). In the present study, a corresponding high level of genetic diversity was observed in all of the examined populations when considering varied gender proportions. The present results are in agreement with previous T. baccata genetic studies employing a variety of genetic marker systems, including nuclear microsatellites, RAPDs (Random Amplified Polymorphic DNA), AFLPs (Amplified Fragment Length Polymorphism), and isozymes (Lewandowski et al. 1995; Hilfiker et al. 2004b; Myking et al. 2009; Zarek 2009; Dubreuil et al. 2010; González-Martínez et al. 2010; Klumpp and Dhar 2011; Trӧber and Ballian 2011; Chybicki et al. 2011; 2012). The results are also in accordance with those reported for other outcrossing, wind-pollinated, conifer forest tree species characterized by a high level of genetic diversity and a low to moderate level of genetic differentiation between populations (Hamrick et al. 1992; Petit et al. 2005). Similar trends to those of T. baccata at the level of genetic variation within and among populations have been described for the long-lived, dioecious common juniper (Juniperus communis L.). Utilizing three polymorphic microsatellite nuclear loci, genetic analysis of nineteen J. communis populations originating from twelve regions of Ireland indicated a high level of heterozygosity, ranging from 0.406 to 0.765 (Provan et al. 2008).
Statistical analyses of the set of estimators describing the genetic structure of male and female T. baccata individuals revealed that there were no statistically significant differences between the two groups. Cao et al. (2004) previously studied genetic variation in T. baccata populations in Germany using isoenzyme markers and compared the genetic structure in male and female individuals. Their analysis revealed minimal significant differences in the values of the tested parameters (He, Ho, A/L), which they suggested to be due to the different number of male and female individuals in the populations and the influence of genetic drift on allele frequencies in the populations. Additionally, Nosrati et al. (2012) measured levels of genetic diversity for male and female individuals of the dioecious atlantic pistachio (Pistacia atlantica Defs.) using RAPD markers. Their study showed that males exhibited a greater level of genetic variation than females. In contrast, Ukwubile et al. (2014) recently determined the genetic characterization of several willow (Salix spp.) genotypes using both RAPDs and nSSRs. They suggested that a set of parameters describing genetic variation revealed no differences between the individuals of Salix spp. grouped according to sex.
A growing number of studies have relied on the identification of outlier loci with distinctly different Fst from those with neutral expectations (Beaumont and Nichols 1996; Vitalis et al. 2001; Beaumont and Balding 2004) as evidence for the presence of non-neutral microsatellite loci in plants (Acheré et al. 2005; Lazrek et al. 2009; Shi et al. 2011). Some nSSRs may be linked to adaptively crucial genes (e.g., sex-linked genes in dioecious species), and their neutral character should be confirmed for the investigation of natural genetic processes leading to genetic depletion (Beaumont and Nicholas 1996; Ohsawa and Ide 2008). Indeed, Cherif et al. (2013) distinguished three potentially sex-linked nSSR loci located in noncoding regions, which are reliable markers of date palm sex. They showed significantly higher genetic differentiation between sexes than other nSSR loci used in their study. These sex-linked loci are heterozygous only in date palm males, and they significantly differed from Hardy-Weinberg (H-W) equilibrium. In contrast, females showed no significant deviation from the H-W expectation. Male-specific alleles allowed the authors to identify the gender in 100% of date palm individuals. Nevertheless, the analysis in the present study confirmed the neutrality of microsatellite nuclear loci originally identified in T. baccata using two different methods: a calescent-based simulation approach performed in Lositian and a hierarchical Bayesian simulation implemented in Bayescan. Because no differences were observed in the level of genetic diversity and genetic differentiation between male and female individuals, the absence of a significant difference indirectly suggests that the tested loci are not sex-linked genetic markers.
There are frequent significant deviations from the equality of sex ratios among natural dioecious plant populations (see review Sinclair et al. 2012). Evidently, unbalanced sex proportions in natural populations have been presumed to affect the pattern of genetic variation in future generations due to genetic drift and increased inbreeding because of non-random mating (see review Sinclair et al. 2012), especially in a case of outcrossing, long-lived dioecious species with repeated episodes of reproduction, such as T. baccata (Iszkuło 2009; Chybicki et al. 2011). Furthermore, the genetic mechanism of sex determination can be labile systems. Environmental factors that determine a fixed sex expression switch on specific genes (Korpelainen 1998). Several authors have reported that an individual T. baccata tree can change its sex (Hartzell 1991), and cosexual yew trees carrying both female and male reproductive structures exist (1%) in the Caucasus Mountains (Pridnya 1984). Furthermore, a low level of genetic variation has been reported for small-female-biased T. baccata populations in Switzerland using RAPD markers (Hilfiker et al. 2004a). These findings indicated that such populations have undergone a higher degree of genetic drift than large populations. Another investigation reported that genotypic diversity of dioecious clonal forest herb dog mercury (Mercurialis perennis) decreased with male-biased sex ratios (Vandepitte et al. 2010). Therefore, the sampling design of collected material should be taken into consideration, as the proportion of male and female individuals in a given population may change due to many environmental factors and stochastic events related to small and isolated populations. The present study was also performed using T. baccata individuals derived from populations where sex ratios deviate from the 1:1 equilibrium expectation, containing between 32–68% females in the population. The number of individuals of opposite sex subjected to analyses also varied in the different populations. However, it was demonstrated that ignoring sex ratios in T. baccata populations had no impact on the assessment of their genetic structure because no correlation was observed between sex ratios and genetic variation parameters or any statistically significant differences in the level of genetic diversity between male and female individuals.
In spite of the fact that the differences at the level of genetic variation between male and female yews are unexpected, this hypothesis was confirmed using microsatellites. The results of the present study are relevant to the practical application of molecular markers in the conservation management of T. baccata genetic resources. The results also demonstrate that the consideration of the T. baccata population sex ratio is not essential to develop a clear understanding of the level of genetic differentiation and diversity within and between populations. It would be very helpful to identify sex-specific for T. baccata markers because there is currently no way to identify the sex of each shrub before reproductive age, and the sex-determining mechanism is still unclear.
Acknowledgements
This research was funded by The State National Forest Holding “Selection of genotypes and populations of English yew for conservation of genetic diversity” and by The Institute of Dendrology Polish Academy of Science. We also acknowledge studies that are part of the program, “Restoration of English yew in Poland”. The authors would also like to thank editor and two anonymous reviewers for making valuable suggestions that improved the manuscript and M. Ratajczak for providing expert technical support.
References
Acheré V., Favre J.M., Besnard G., Jeandroz S. (2005). Genomic organization of molecular differentiation in Norway spruce (Picea abies). Molecular Ecology 14: 3191–3201. http://dx.doi.org/10.1111/j.1365-294X.2005.02646.x.
Antao T., Lopes A., Lopes R.J., Beja-Pereira A., Luikart G. (2008). LOSITAN: a workbench to detect molecular adaptation based on Fst-outlier method. BMC Bioinformatics 9: 323. http://dx.doi.org/10.1186/1471-2105-9-323.
Barrett S.C.H., Hough J. (2013). Sexual dimorphism in flowering plants. Journal of Experimental Botany 64: 67–82. http://dx.doi.org/10.1093/jxb/ers308.
Beaumont M.A., Balding D.J. (2004). Identification adaptive genetic divergence among populations from genome scans. Molecular Ecology 13: 969–980. http://dx.doi.org/10.1111/j.1365-294X.2004.02125.x.
Beaumont M.A., Nicholas R.A. (1996). Evaluating loci for use in the genetic analysis of population structure. Proceeding of the Royal Society, Biological Sciences 263: 1619–1623. http://dx.doi.org/10.1098/rspb.1996.0237.
Cao C.P., Leinemann L., Ziehe M., Finkeldey R. (2004). Study of genetic variation and differentiation of yew (Taxus baccata L.) stands using izoenzyme and DNA marker. Allgemeine Forest und Jagdzeitung 175: 21–28.
Cedro A., Iszkuło G. (2011) Do females differ from males of European yew (Taxus baccata L.) in dendrochronological analysis? Tree-Ring Research 67: 3–11. http://dx.doi.org/10.3959/2009-9.1.
Chapuis M.P., Estoup A. (2007). Microsatellite null alleles and estimation of population differentiation. Molecular Biology and Evolution 24: 621–631. http://dx.doi.org/10.1093/molbev/msl191.
Cherif E., Zehdi S., Castillo K., Chabrillange N., Abdoulkader S., Pintaud J.C., Santoni S., Salhi-Hannachi A., Glémin S., Aberlenc-Bertossi F. (2013). Male-specific DNA markers provide genetic evidence of an XY chromosome system, a recombination arrest and allow the tracing of paternal lineages in date palm. New Phytologist 197(2): 409–15. http://dx.doi.org/10.1111/nph.12069.
Chybicki I.J., Burczyk J. (2009). Simultaneous estimation of null alleles and inbreeding coefficients. Journal of Heredity 100: 106–113. http://dx.doi.org/10.1093/jhered/esn088.
Chybicki I.J., Oleksa A., Burczyk J. (2011). Increased inbreeding and strong kinship structure in Taxus baccata estimated from both AFLP and SSR data. Heredity 107: 589–600. http://dx.doi.org/10.1038/hdy.2011.51.
Chybicki I.J., Oleksa A., Kowalkowska K. (2012). Variable rates of random drift in protected populations of English yew: implications for gene pool conservation. Conservation Genetics 13: 899–911. http://dx.doi.org/10.1007/s10592-012-0339-9.
Dubreuil M., Sebastiani F., Mayol M., González-Martínez S., Riba M., Vendramin G. (2008). Isolation and characterization of polymorphic nuclear microsatellite loci in Taxus baccata L. Conservation Genetic 9: 1665–1668. http://dx.doi.org/10.1007/s10592-008-9515-3.
Dubreuil M., Riba M., Gonzalez-Martinez S., Vendramin G.G., Sebastiani F., Mayol M. (2010). Genetic effects of chronic habitat fragmentation revisited: strong genetic structure in temperate tree, Taxus baccata (Taxaceae), with great dispersal capability. American Journal of Botany 97: 303–310. http://dx.doi.org/10.3732/ajb.0900148.
Dumolin S., Demesure B., Petit R.J. (1995). Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theoretical and Applied Genetics 91: 1253–1256. http://dx.doi.org/10.1007/BF00220937.
Engen S., Lande R., Saether B.E. (2003). Demographic stochasticity and allee effects in populations’ with two sexes. Ecology 84: 2378–2386. http://dx.doi.org/10.1890/02-0123.
Eppley S.M. (2005). Spatial segregation of the sexes and nutrients affect reproductive success in a dioecious wind-pollinated grass. Plant Ecology 181: 179–190. http://dx.doi.org/10.1007/s11258-005-6142-7.
Excoffier L., Laval G., Schneider S. (2005). Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics 1: 47–50.
Fisher R.A. (1930). The genetical theory of natural selection. Clarendon Press, Oxford.
Freeman D.C., Doust J.L., El-Keblawy A., Miglia K.J., McArthur E.D. (1997). Sexual specialization and inbreeding avoidance in the evolution of dioecy. Botanical Review 63(1): 65–92. http://dx.doi.org/10.1007/BF02857918.
Foll M., Gaggiotti O. (2008). A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180: 977–993. http://dx.doi.org/10.1534/genetics.108.092221.
Givnish T.J. (1980). Ecological constraints on the evolution of breeding systems in seed plants: dioecy and dispersal in gymnosperms. Evolution 34: 959–972.
González-Martínez S.C., Dubreuil M., Riba M., Vendramin G.G., Sebastiani F., Mayol M. (2010). Spatial genetic structure of Taxus baccata L. in the western Mediterranean Basin: past and present limits to gene movement over a broad geographic scale. Molecular Phylogenetics and Evolution 55: 805–815. http://dx.doi.org/10.1016/j.ympev.2010.03.001.
Goudet J. (2001). FSTAT, a program to estimate and test gene diversities and fixation indices. Version 2.9.3. http://www2.unil.ch/popgen/softwares/fstat.htm. [Cited 15 May 2013].
Hamrick J.L., Godt M.J.W., Sherman-Broyles S.L. (1992). Factors influencing levels of genetic diversity in woody plant species. New Forests 6: 95–124. http://dx.doi.org/10.1007/BF00120641.
Hartzell H.R. Jr. (1991). The yew tree: a thousand whispers. Hulogosi, Eugene, Oregon, USA.
Hedrick P.W. (2000). Genetics of populations. Jones and Bartlett Publisher, Inc., Boston, MA.
Heilbuth J.C. (2000). Lower species richness in dioecious clades. The American Naturalist 156: 241–261. http://dx.doi.org/10.1086/303389.
Hilfiker K., Holderegger R., Rotach P., Gugerli F. (2004a). Dynamics of genetic variation in Taxus baccata: local versus regional perspectives. Canadian Journal of Botany 82: 219–227. http://dx.doi.org/10.1139/B03-136.
Hilfiker K., Gugerli F., Schutz J.P., Rotach P., Holderegger R. (2004b). Low RAPD variation and female-biased sex ratio indicate genetic drift in small populations of the dioecious conifer Taxus baccata in Switzerland. Conservation Genetics 5: 357–365. http://dx.doi.org/10.1023/B:COGE.0000031144.95293.1b.
Iszkuło G., Jasińska A., Giertych M., Boratyński A. (2009). Do secondary sexual dimorphism and female intolerance to drought influence the sex ratio and extinction risk of Taxus baccata? Plant Ecology 200: 229–240. http://dx.doi.org/10.1007/sl1258-008-9447-5.
Jeffreys H. (1961). The theory of probability. (3rd ed.). Clarendon Press, Oxford. p. 432.
Klumpp R., Dhar A. (2011). Genetic variation of Taxus baccata L. populations in the Eastern Alps and its implications for conservation management. Scandinavian Journal of Forest Research 26: 294–304. http://dx.doi.org/10.80/02827581.2011.566888.
Korpelainen H. (1998). Labile sex expression in plants. Biological Review 73: 157–180. http://dx.doi.org/10.1111/j.1469-185X.1997.tb00028.x.
Lazrek F., Roussel V., Ronofort J., Cardient G., Chardon F. (2009). The use of neutral and non-neutral SSRs to analyze the genetic structure of Tunisian collection of Medicago truncatula lines and to reveal associations with ecoenvironmental variables. Genetica 135: 391–402. http://dx.doi.org/10.1007/s10709-008-9285-3.
Lewandowski A., Burczyk J., Mejnartowicz L. (1995). Genetic structure of English yew (Taxus baccata L.) in the Wierzchlas Reserve: implications for genetic conservation. Forest Ecology and Management 73: 221–227. http://dx.doi.org/10.1016/0378-1127(94)0377-E.
Litkowiec M., Plitta B.P., Lewandowski A. (2013). Importance of genetic variation for conservation of English yew genetic resources in Europe. [Znaczenie zmienności genetycznej dla ochrony zasobów genowych cisa pospolitego w Europie]. Sylwan 157: 754–760. [Abstract and summary in English].
Mercer C.A., Eppley S.M. (2010). Intersexual competition in a dioecious grass. Ocologia 164: 657–664. http://dx.doi.org/10.1007/S00442-010-1675-4.
Myking T., Vakkari P., Skroppa T. (2009). Genetic variation in northern marginal Taxus baccata L. populations. Implications for conservation. Forestry 82: 529–539. http://dx.doi.org/10.1093/forestry/cpp022.
Nosrati H., Ali Husainpourfeizi M., Khorasani M., Razban-Haghighi A., Nikniazi M. (2012). Sex ratio and genetic diversity in the dioecious Pistacia atlantica (Anacardiaceae). Journal of Agrobiology 29: 41–46. http://dx.doi.org/10.2478/v10146-012-0006-2.
Ohsawa T., Ide Y. (2008). Global patterns of genetic variation in plant species along vertical and horizontal gradients on mountains. Global Ecology and Biogeography 17: 152–163. http://dx.doi.org/10.1111/j.1466-8238.2007.00357.x.
Pannell J., Barrett S.C.H. (1998). Baker’s law revisited: reproductive assurance in a metapopulation. Evolution 52: 657–668.
Peakall R., Smouse P.E. (2006). GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6: 288–295. http://dx.doi.org/10.1111/j.1471-8286.2005.01155.x.
Petit R.J., Duminil J., Fineschi S., Hampe A., Salvani D., Vendramin G.G. (2005). Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Molecular Ecology 14: 689–701. http://dx.doi.org/10.1111/j.1365-294X.2004.02410.x.
Pridnya M.V. (1984). Phytocoenotic status and structure of the Khosta common-yew population in the Caucasus Biosphere Reserve. Soviet Journal of Ecology 15: 1–6.
Provan J., Beatty G.E., Hunter M.A., McDonald R.A., McLaughlin E., Preston S.J., Wilson S. (2008). Restricted gene flow in fragmented populations of wind-pollinated tree. Conservation Genetic 9: 1521–1532. http://dx.doi.org/10.1007/s10592-007-9484-y.
Renner S.S., Ricklefs R.E. (1995). Dioecy and its correlates in the flowering plants. American Journal of Botany 82: 596–606. http://dx.doi.org/10.2307/2445418.
Shi M-M., Michalski S.G., Chen X-Y., Durka W. (2011). Isolation by elevation: genetic structure at neutral and putatively non-neutral loci in a dominant tree of subtropical forests, Castanopsis eyrei. PLose ONE 6: e21302. http://dx.doi.org/10.1371/journal.pone.0021302.
Sinclair J.P., Emlen J., Freeman D.C. (2012). Biased sex ratios in plants: theory and trends. Botanical Review 78: 63–86. http://dx.doi.org/10.1007/s12229-011-9065-0.
Soza V.L., Brunet J., Liston A., Smith P.S., Di Stilio V.S. (2012). Phylogenetic insights into the correlates of dioecy in meadow-rues (Thalictrum, Ranunculaceae). Molecular Phylogenetics and Evolution 63: 180–192. http://dx.doi.org/10.1016/j.ympev.2012.01.009.
Svenning J.CH., Magård E. (1999). Population Ecology and conservation status of the last natural population of English yew (Taxus baccata) in Denmark. Biological Conservation 88: 173–182. http://dx.doi.org/10.1016/S0006-3207(98)00106-2.
Thomas P.A., Polwart A. (2003). Taxus baccata L. Journal of Ecology 91: 489–524. http://dx.doi.org/10.1046/j.1365-2745.2003.00783.x.
Trӧber U., Ballian D. (2011). Genetic characterization of English yew (Taxus baccata L.) populations in Bosnia and Herzegovina. European Journal of Forest Research 130: 479–489. http://dx.doi.org/10.1007/s10342-010-0436-6.
Ukwubile C.A., Ahuchaogu CH.E., Tajudeen B.L. (2014). Congruence of random amplification of polymorphic deoxyribonucleic acid (RAPD) and simple sequence repeats (SSR) markers in genetic characterization of willow (Salix spp.). European Journal of Biotechnology and Bioscience 2: 11–23.
Vamosi J.C., Vamosi S.M. (2005). Present day risk of extinction may exacerbate the lower species richness of dioecious clades. Diversity and Distributions 11: 25–32. http://dx.doi.org/10.1111/j.1366-9516.2005.00119.x.
Vandepitte K., Roldán-Ruiz I., Honnay O. (2009). Reproductive consequences of mate quantity versus mate diversity in a wind-pollinated plant. Acta Oecolociga 35: 548–553. http://dx.doi.org/10.1016/j.actao.2009.04.004.
Vandepitte K., Honnay O., De Meyer T., Jacquemyn H., Roldán-Ruiz I. (2010). Patterns of sex ratio variation and genetic diversity in the dioecious forest perennial Mercurialis perennis. Plant Ecology 206: 105–114. http://dx.doi.org/10.1007/s11258-009-9627-y.
Vitalis R., Dawson K., Bourost P. (2001). Interpretation of variation across marker loci as evidence of selection. Genetics 158: 1811–1823.
Weir B., Cockerham C. (1984). Estimating F-Statistics for the analysis of population structure. Evolution 38: 1358–1370. http://dx.doi.org/10.2307/2408641.
Zarek M. (2009). RAPD Analysis of genetic structure in four natural populations of Taxus baccata from southern Poland. Acta Biologica Cracoviensia Series Botanica 51: 67–75.
Total of 60 references

