Evidences of genetic bottleneck and fitness decline in Luehea divaricata populations from southern Brazil
Stefenon V. M., Nagel J. C., Poletto I. (2016). Evidences of genetic bottleneck and fitness decline in Luehea divaricata populations from southern Brazil. Silva Fennica vol. 50 no. 5 article id 1566. https://doi.org/10.14214/sf.1566
Highlights
- Signatures of genetic bottlenecks and reduction of populations’ fitness were observed in populations of Luehea divaricata in southern Brazil
- Lower levels of observed heterozygosity are correlated with populations’ fitness, decreasing germination capacity and increasing the proportion of anomalous germinated plantlets
- Promoting the connection among populations is proposed as a key strategy towards conservation of L. divaricata genetic resources in its southernmost distribution range.
Abstract
Extant populations growing in regions that were refugia during the last glacial period are expected to show higher genetic diversity than populations that moved from these refugia into new areas in higher latitudes. Such new populations likely faced harsher climatic conditions, being established with reduced population size and experiencing the effects of genetic bottlenecks. In this study we employed data from nuclear SSR markers for detecting molecular signatures of genetic bottlenecks, and germination experiments to evaluate reduction of populations’ fitness in natural populations of Luehea divaricata Mart. et Zucc., growing in the southern range of the species distribution (around 30°S latitude). Signatures of genetic bottlenecks and reduction of populations’ fitness were observed in all populations. Lower levels of observed heterozygosity are correlated with populations’ fitness, decreasing germination capacity and increasing the proportion of anomalous germinated plantlets. Promoting the connection among populations is proposed as a key strategy towards conservation of L. divaricata genetic resources in its southernmost distribution range. The offspring from crosses among populations would significantly increase the observed heterozygosity and fitness of multiple populations.
Keywords
demographic history;
effective population size;
genetic resources conservation;
Pampa biome
-
Stefenon,
Nucleus of Genomics and Molecular Ecology, Interdisciplinary Center of Biotechnological Research, Universidade Federal do Pampa, BR290 km, 97300-000, São Gabriel, RS, Brazil
E-mail
valdirstefenon@unipampa.edu.br

- Nagel, Nucleus of Genomics and Molecular Ecology, Interdisciplinary Center of Biotechnological Research, Universidade Federal do Pampa, BR290 km, 97300-000, São Gabriel, RS, Brazil E-mail jordana.nagel@yahoo.com.br
- Poletto, Laboratory of Plant Protection, Universidade Federal do Pampa, BR290 km, 97300-000, São Gabriel, RS, Brazil E-mail igorpoletto@unipampa.edu.br
Received 11 February 2016 Accepted 10 October 2016 Published 9 December 2016
Views 120311
Available at https://doi.org/10.14214/sf.1566 | Download PDF

1 Introduction
About 65 million years ago, during the Tertiary, the Earth’s climate became colder, with recurrent oscillations that increased in amplitude and lead to the series of major ice ages of the Quaternary (Hewitt 2000). Present patterns of genetic diversity and variation observed in natural populations of the temperate zones have been postulated to be result of the last glacial period and the subsequent recolonization when animal and plant species expanded into lands earlier not able to be inhabited (Dumolin-Lanpègue et al. 1997). Similar movements of animals and plants may have occurred into subtropical and temperate zones of the Southern Hemisphere. During major glaciations the polar ice sheets spread considerably, and vegetation zones were compressed basically towards the Equator (Hewitt 2000). When climatic conditions became more favorable during the present interglacial, some species that survived in these refugia moved into new areas in higher latitudes. Thus, extant populations growing in higher latitudes likely faced harsher climatic conditions in comparison to populations from Equatorial zones, being established with reduced effective population size.
Reduction of population census size (demographic bottlenecks) and reduction of effective population size (genetic bottlenecks) as result of climatic shifts, habitat fragmentation and populations’ isolation have been documented as a recurrent event in forest tree populations (e.g. Ledig et al. 1999; Al-Rabab’ah and Williams 2004; Stefenon et al. 2008). Such reductions of effective population size may result in very negative outcomes to these populations, guiding them to a decline in adaptive and reproductive potentials, and to increased risk of extinction (Nei et al. 1975) as effect of demographic, genetic, and environmental stochasticity (Luijten et al 2000). A large number of studies have shown that the postglacially colonized regions have lower genetic diversity in comparison to refugia areas (Hewitt 2000).
Although demographic and genetic bottlenecks may be directly correlated, populations experiencing a reduction in census size may not suffer a severe effective size reduction, as well as currently large populations may have experienced drastic genetic bottlenecks in a recent past (Peery et al. 2012). Thus, management and conservation programs for threatened, ecologically important or commercially valuable plant species should include genetic monitoring, as reduction in population effective size can occur without being detected by traditional demographic monitoring approaches. For instance, due to the high reproductive output of forest plant species, there is always the chance that the next generation is descended from a small number of individuals, resulting in a cryptic genetic bottleneck without a reduction of population census size (Luikart et al. 1998a).
Populations with declined effective size may suffer the effects of genetic drift, which will lead to the fixation of alleles and loss of genetic variation (Luijten et al. 2000), increasing genetic differentiation among populations and generating inbred progenies. Such progenies will in turn be composed by high incidence of individuals that are homozygous for alleles identical by descent (Keller and Waller 2002) and suffer the effects of the inbreeding depression. Inbreeding depression negatively affects fitness throughout the life cycle of plants (Finkeldey and Hattemer 2007), reducing the reproductive potential (and thus the adaptive efficiency). Several studies, based mainly on comparing plant populations that differ in size or levels of genetic variation, reveal significant inbreeding effects on seed set, germination, survival and resistance to stress (Keller and Waller 2002).
In a recent study (Nagel et al. 2015), significant inbreeding coefficient and high levels of putative half- and full-sibs were recorded in five populations of Luehea divaricata Mart. et Zucc. (Malvaceae) from Southern Brazil. Since the census population sizes in widespread tree species tend to be historically large, while effective population sizes tend to be only slightly lower than census numbers (Al-Rabab’ah and Williams 2004), these genetic patterns may be indicatives of an ancient founder effect (Nagel et al. 2015).
Luehea divaricata occurs through out a large range of geographic distribution in South America (Fig. 1), growing mainly along rivers in riparian forests on both wet and drained soils, in semi-devastated forests, in secondary forests and coppices (Reitz et al. 1988). It is a cosexual hermaphrodite species pollinated mainly by bees and hummingbirds (Backes and Irgang 2002) and with wind dispersed seeds (Paoli 1995). This species can reach up to 25 m in height and 80 cm diameter at breast hight (DBH) and is frequently recommended in reforestation programs of riparian forests in Brazil (Ceconi et al. 2006). Due to the high flexibility of its wood, L. divaricata is also employed as raw material for handmade furniture. Leaves of L. divaricata are used as phytotherapic product for dysentery, leucorrhea, rheumatism, blennorrhoea and tumors, while their roots have depurative properties (Tanaka et al. 2005) and the infusion of its flowers and stem bark is used in the treatment of respiratory diseases (Teixeira et al. 2016).
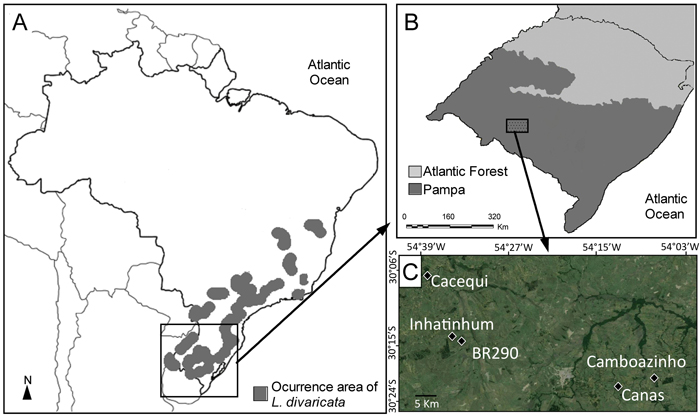
Fig. 1. (A) Distribution area of L. divaricata in Southern America, including Brazil and small border areas in Argentina, Uruguay and Paraguay (adapted from Carvalho 2001). (B) Location of the Pampa biome in southern Brazil. (C) Location of the five studied populations within the Pampa biome.
Although L. divaricata is currently not listed as a threatened species in official documents, the fragmentation of its populations may lead the species to endangerment (Conson et al. 2013; Nagel et al. 2015). In such conditions, the maintenance of species adaptability is not assured, given the negative effects of genetic drift and inbreeding in isolated populations, mainly in fragments with small effective population size (Nagel et al. 2015).
When analyzed in the appropriate theoretical framework, molecular genetic methods provide a powerful tool for inferring the demographic history of populations and recent reductions in effective population size (Cornuet and Luikart 1996; Luikart et al. 1998b; Beaumont 1999; Wang 2005). Reductions in effective population sizes are expected to directly affect the distribution of alleles in neutral loci (Nei et al. 1979). Using neutral markers such as microsatellites, statistical tests can detect signatures of recent severe effective population size reductions, such as drifts in the expected distribution of alleles based on their frequency (Luikart et al. 1998b), heterozygosity excess relative to expected under mutation-drift equilibrium (Cornuet and Luikart 1996), low rate of the alleles number in relation to the overall range in fragment sizes (M-ratio; Garza and Williamson 2001) and increased linkage disequilibrium (Flint-Garcia et al. 2003).
In this study, we used different tests based on genetic data from microsatellite markers to examine whether populations of L. divaricata from the Brazilian Pampa exhibit molecular signatures of historical genetic bottlenecks. In addition we estimated the mean reproductive success of these populations by measuring seed germination traits as an indirect estimation of the populations’ fitness. Correlation analysis was employed to evaluate the influence of inbreeding depression and germination traits. We intended to answer the following questions: 1) Have populations of L. divaricata from their southern distribution range of distribution experienced genetic bottleneck events? 2) If so, have these events caused loss of population fitness due to inbreeding depression?
2 Material and methods
2.1 Sampling strategy and Microsatellite genotyping
Plant material of L. divaricata was sampled in five natural populations in southern Brazil, within the Brazilian Pampa (Fig. 1), a region dominated by a savanna-like environment (Roesch et al. 2009). Healthy leaves of at least 30 adult individuals were collected in five populations of L. divaricata (n = 167) and genotyped using nuclear microsatellite markers. Given the small extent of the studied forest fragments, this sample size covered at least 90% of the adult individuals in each population. The five populations are separated from one another by grassland formations, croplands or urban areas. About 50 mg of plant material was washed with 70% ethanol and distilled water, disrupted in collection microtubes using the TissueLyser (Qiagen®) grinder with 3 mm tungsten beads, and total DNA was extracted using the Invisorb Plant Mini Kit (Invitek®), following the instructions of the manufacturer. Isolated DNA was eluted in 100 μL of elution buffer and deposited at –20 °C until use. The quality and the amount of the extracted DNA were evaluated on a NanoVueTM Plus Spectrophotometer (GE Healthcare).
Ten microsatellite loci developed by Ruas et al. (2009) were tested, and genotypes of all samples were scored at five putatively neutral nuclear microsatellite loci (Ldiv31, Ldiv40, Ldiv48A, Ldiv55 and Ldiv58), which revealed reliable banding patterns in the gels. The PCR amplification of all loci was performed in an Eppendorf® Thermal Cycler in 25 μL reactions, comprising 40 ng of template DNA, 1X PCR buffer, 2.5 mM MgCl2, 1.0 U Taq DNA polymerase (Invitrogen®), 0.2 mM of each dNTP and 10 μM of each primer. PCR parameters were defined as described by Ruas et al. (2009), concerning one initial cycle at 94 °C for 4 min, followed by 16 touchdown cycles of 94 °C for 30 s, 65 °C for 30 s (decreasing of 1 °C per cycle) and 72 °C for 30 s, continuing with 20 additional cycles of 94 °C for 30 s, 50 °C for 30 s, 72 °C for 30 s and a final extension step at 72 °C for 7 min. All fragments were separated on 30 cm 10% polyacrylamide gels, stained with silver nitrate as described by Stefenon and Nodari (2003) and photographed using a digital camera. Alleles were scored from the digital pictures using the software TotalLabTM TL120 (Nonlinear Dynamics).
2.2 Search for signatures of reduced effective population size
Signature of reduced population size was tested using different approaches, aiming to assess signatures of genetic bottleneck events over a wide variety of demographic scenarios (different pre-bottleneck θ, bottleneck severity, time since the bottleneck beginning and speed of sample size recover) and distinct mutation models, the stepwise mutation model (SMM) or the two-phased mutation model (TPM).
The first method consists in testing for heterozygosity excess in comparison to predictable heterozygosity under mutation-drift equilibrium considering the observed number of alleles (Cornuet and Luikart 1996). Estimates of the expected heterozygosity under mutation-drift equilibrium (HEQ) were obtained by simulating the coalescent process (Kingman 1982) using 10 000 iterations for each population following the stepwise mutation model (SMM; Ohta and Kimura 1973) and the two-phased mutation model (TPM; Di Rienzo et al. 1994), as these are considered most appropriate for microsatellites (Di Rienzo et al. 1994; Garza and Williamson 2001). The Hardy-Weinberg heterozygosity (HE) is based on the allelic frequencies and was computed as ![]() where pi is the frequency of the ith allele. Bottlenecked populations are expected to reveal a significant excess of Hardy-Weinberg heterozygosity (HE) in comparison to the heterozygosity estimated under mutation-drift equilibrium (HEQ). Statistical significance of the analyses was assessed through the one-tail Wilcoxon test (Luikart et al. 1998a). All computations were performed using the software Bottleneck 1.2.02 (Piry et al. 1999).
where pi is the frequency of the ith allele. Bottlenecked populations are expected to reveal a significant excess of Hardy-Weinberg heterozygosity (HE) in comparison to the heterozygosity estimated under mutation-drift equilibrium (HEQ). Statistical significance of the analyses was assessed through the one-tail Wilcoxon test (Luikart et al. 1998a). All computations were performed using the software Bottleneck 1.2.02 (Piry et al. 1999).
The second method assumes that populations that experienced a recent reduction in effective size tend to show a shift in the allele frequency distribution, with the incidence of alleles at frequency inferior to 0.1 becoming lower than the incidence of alleles in intermediate allele frequency classes (Luikart et al. 1998b). For this mode-shift analysis under the stepwise mutation model, alleles were grouped into ten allele frequency classes (0.001–0.100, 0.101–0.200, 0.201–0.300, etc.) and plotted into a frequency histogram, as performed in the software Bottleneck 1.2.02 (Piry et al. 1999). Bottlenecked populations tend to present a shifted mode distribution, while populations that did not suffer reduction in effective size display a L-shaped distribution.
The third method comprises the analysis of the ratio of the total number of alleles (k) to the overall range in allele size (r) as ![]() (Garza and Williamson 2001). To test whether M-values are lower than expected (cases in which a population experienced a bottleneck), the critical M-value (Mc) was estimated for each population by simulating 10 000 replicates of the coalescent process conditional on the populations’ allelic information. We used the two-phased mutation model assuming a proportion of one-step mutations (ps) of 0.22, a mean size of non one-step mutations (Δg) of 3.1 (following Vigoroux et al. 2002 and Peery et al. 2012) and pre-bottleneck effective population sizes Ne = 500, Ne = 100 and Ne = 50, considering a mutation rate μ = 5×10–4. The analysis was performed using the software M_P_val (Garza and Williamson 2001).
(Garza and Williamson 2001). To test whether M-values are lower than expected (cases in which a population experienced a bottleneck), the critical M-value (Mc) was estimated for each population by simulating 10 000 replicates of the coalescent process conditional on the populations’ allelic information. We used the two-phased mutation model assuming a proportion of one-step mutations (ps) of 0.22, a mean size of non one-step mutations (Δg) of 3.1 (following Vigoroux et al. 2002 and Peery et al. 2012) and pre-bottleneck effective population sizes Ne = 500, Ne = 100 and Ne = 50, considering a mutation rate μ = 5×10–4. The analysis was performed using the software M_P_val (Garza and Williamson 2001).
Since a genetic bottleneck may also generate linkage disequilibrium in natural populations (Wang et al 1998; Flint-Garcia et al. 2003), the nonrandom association of alleles at different loci was also investigated. Contingency tables were created for all pairs of loci in each population and the log likelihood ratio statistic (G-test) was computed for each table using the Markov chain algorithm with 10 000 dememorisation steps, 1000 batches and 10 000 iterations per batch in the software GenePop (Raymond and Rousset 1995).
2.3 Estimation of populations’ reproductive success
The mean population reproductive success was evaluated through the analysis of seeds’ germinability, the speed of germination and the proportion of anomalous germinated plantlets. Open-pollinated seeds were collected from six mother trees (25 seeds/tree) in each population, except in population BR290 that failed in producing flowers and fruits during the spring season in year 2014. All 600 seeds were placed in petri dishes lined with filter paper, saturated with distilled water, and placed in a growth chamber at 25 ± 1 °C and a photoperiod of 12 h of light to germinate. Embryos emergence was monitored each seven days during eight weeks.
Seeds’ germinability was estimated as the percentage of seeds in which the germination process reached the stage of emergence of a live embryo (Ranal and Santana 2006). The speed of germination (SG) was estimated following Maguire (1962) based on the summation of the emerged embryos at each counting time as ![]() where E1 is the number of emerged embryos at the first week, t1 is the time of the first counting (1st week) and n is the last counting time (8th week). The proportion of anomalous germinated plantlets was determined as the percentage of germinated plantlets that presented atypical development as the absence of one or both cotyledons, absence of the root systems or distorted growth of the plantlet.
where E1 is the number of emerged embryos at the first week, t1 is the time of the first counting (1st week) and n is the last counting time (8th week). The proportion of anomalous germinated plantlets was determined as the percentage of germinated plantlets that presented atypical development as the absence of one or both cotyledons, absence of the root systems or distorted growth of the plantlet.
The correlation between each germination trait and the inbreeding coefficient (f) and the observed heterozygosity (HO) was estimated through the Pearson’s product-moment correlation coefficient (r) using the Free Statistics Software v1.1.23-r7 (Wessa 2016). The inbreeding coefficient is an estimation of the level of genetic correlation between individuals within the population, and was measured as the kinship coefficient between gene copies within individuals (Nagel et al. 2015), while the observed heterozygosity is an estimation of the actual population’s level of genetic diversity.
3 Results
3.1 Heterozygosity excess
Signature of recent genetic bottleneck was detected in the analysis of heterozygosity excess for populations Camboazinho, Canas and BR290 for the TPM model and just in population Camboazinho for the SSM model, with statistically significant difference between HE and HEQ (p < 0.05; Table 1). The mean expected heterozygosity under Hardy-Weinberg expectations ranged from He = 0.53 in population Cacequi to He = 0.77 in population Canas (Table 1).
| Table 1. Heterozygosity excess, mode-shift distribution and M-ratio analyses of bottleneck signature for populations of L. divaricata, based on five microsatellite loci. Values in bold means positive signature of a genetic bottleneck event. | |||||||||
| Population | Genetic diversity parameters | Loci with heterozygosity excess | Mode-shift distribution | M-ratio analysis | |||||
| k | HE | TPM | SMM | M-value | Ne = 500 | Ne = 100 | Ne = 50 | ||
| Camboazinho | 4.4 | 0.69 | 5 (p = 0.02) | 5 (p = 0.02) | L-shaped | 0.61 | p = 0.030 | p = 0.009 | p = 0.007 |
| Canas | 7.0 | 0.77 | 5 (p = 0.02) | 4 (p = 0.11) | L-shaped | 0.64 | p = 0.047 | p = 0.015 | p = 0.012 |
| Inhatinhum | 4.8 | 0.61 | 4 (p = 0.31) | 3 (p = 0.41) | L-shaped | 0.63 | p = 0.045 | p = 0.015 | p = 0.009 |
| Cacequi | 3.8 | 0.53 | 4 (p = 0.11) | 2 (p = 0.68) | L-shaped | 0.46 | p = 0.006 | p = 0.001 | p = 0.002 |
| BR290 | 4.4 | 0.61 | 4 (p = 0.03) | 4 (p = 0.31) | Shifted | 0.51 | p = 0.026 | p = 0.006 | p = 0.002 |
| K: number of alleles; HE: Hardy-Weinberg heterozygosity; TPM: two-phased mutation model; SMM: Stepwise mutation model; M-value: ratio of the total number of alleles to the overall range in allele size; Ne: effective population size | |||||||||
3.2 Allelic Frequencies Distribution
Concerning the distribution of allelic frequencies, just population BR290 revealed a shifted distribution, suggesting the occurrence of recent reduction in population effective size. All other populations presented normal L-shaped distribution of the alleles frequency (Table 1; Fig. 2).
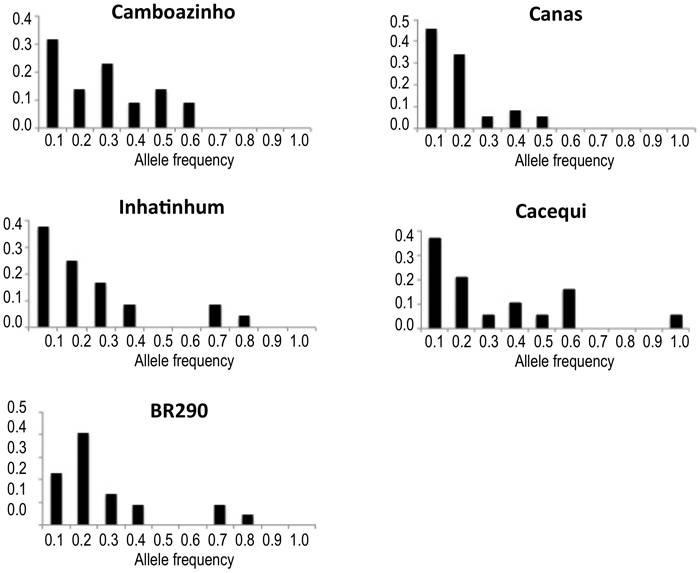
Fig. 2. Plotting of the frequency distribution of allele classes for microsatellite markers. Populations Camboazinho, Canas, Inhatinhum and Cacequi revealed a non-shifted alleles distribution, meaning absence of bottleneck signatures. Population BR290 revealed a shifted distribution, indicating historical reduction of the effective population size.
3.3 -Ratio analysis
For the M-ratio analysis, all populations revealed M-values bellow the threshold estimation M = 0.67 proposed by Garza and Williamson (2001) as an upper limit for post-bottleneck M-values. The lowest M-value was observed in population Cacequi (M = 0.46) and the highest in population Canas (M = 0.64; Table 1). Additional evidences of a recent genetic bottleneck in all populations were observed when considering the critical M-value (Mc) estimated for each population. Independent of the pre-bottleneck effective population size considered (Ne = 50, 100 or 500), the M-values of all populations were significantly lower (p < 0.05; Table 1) than the estimated Mc-values. The mean number of alleles observed in each population ranged from k = 3.8 (population Cacequi) to k = 7.0 (population Canas; Table 1).
3.4 Linkage disequilibrium analysis
Significant linkage disequilibrium (LD) was detected in 14 out of the 50 pairwise loci combinations in the five populations (p < 0.05; Table 2). However, no pairs of loci revealed significant LD in all populations. Population Camboazinho presented five pairs of loci (50%) with significant LD and population Canas presented seven pairs (70%), suggesting effects of genetic bottleneck events. Populations Inhatinhum and BR290 presented one pair each (10%), while no pair of loci presented significant LD in population Cacequi, failing to capture signature of effective population size decline in this analysis.
| Table 2. Linkage disequilibrium analysis of five microsatellite loci in populations of L. divaricata. Values in bold means positive signature of a genetic bottleneck event. | |||
| Pop | Pair of loci | p-value | |
| Camboazinho | Ldiv31 | Ldiv40 | 1.000 |
| Ldiv31 | Ldiv48 | 0.724 | |
| Ldiv40 | Ldiv48 | 0.048 | |
| Ldiv31 | Ldiv55 | 0.684 | |
| Ldiv40 | Ldiv55 | <0.001 | |
| Ldiv48 | Ldiv55 | 0.032 | |
| Ldiv31 | Ldiv58 | 0.314 | |
| Ldiv40 | Ldiv58 | 0.043 | |
| Ldiv48 | Ldiv58 | 0.116 | |
| Ldiv55 | Ldiv58 | 0.001 | |
| Canas | Ldiv31 | Ldiv40 | 0.049 |
| Ldiv31 | Ldiv48 | 0.085 | |
| Ldiv40 | Ldiv48 | 0.302 | |
| Ldiv31 | Ldiv55 | 0.001 | |
| Ldiv40 | Ldiv55 | 0.001 | |
| Ldiv48 | Ldiv55 | <0.001 | |
| Ldiv31 | Ldiv58 | 0.479 | |
| Ldiv40 | Ldiv58 | 0.018 | |
| Ldiv48 | Ldiv58 | 0.001 | |
| Ldiv55 | Ldiv58 | 0.003 | |
| Inhatinhum | Ldiv31 | Ldiv40 | 0.372 |
| Ldiv31 | Ldiv48 | 0.639 | |
| Ldiv40 | Ldiv48 | 0.265 | |
| Ldiv31 | Ldiv55 | 0.381 | |
| Ldiv40 | Ldiv55 | 0.350 | |
| Ldiv48 | Ldiv55 | 0.065 | |
| Ldiv31 | Ldiv58 | 0.850 | |
| Ldiv40 | Ldiv58 | 0.003 | |
| Ldiv48 | Ldiv58 | 0.919 | |
| Ldiv55 | Ldiv58 | 0.105 | |
| Cacequi | Ldiv31 | Ldiv40 | 0.746 |
| Ldiv31 | Ldiv48 | 0.236 | |
| Ldiv40 | Ldiv48 | 0.354 | |
| Ldiv31 | Ldiv55 | 0.161 | |
| Ldiv40 | Ldiv55 | 0.482 | |
| Ldiv48 | Ldiv55 | 1.000 | |
| Ldiv31 | Ldiv58 | 0.789 | |
| Ldiv40 | Ldiv58 | 0.239 | |
| Ldiv48 | Ldiv58 | 0.753 | |
| Ldiv55 | Ldiv58 | 0.966 | |
| BR290 | Ldiv31 | Ldiv40 | <0.001 |
| Ldiv31 | Ldiv48 | 0.407 | |
| Ldiv40 | Ldiv48 | 0.586 | |
| Ldiv31 | Ldiv55 | 0.724 | |
| Ldiv40 | Ldiv55 | 0.097 | |
| Ldiv48 | Ldiv55 | 0.460 | |
| Ldiv31 | Ldiv58 | 0.190 | |
| Ldiv40 | Ldiv58 | 0.143 | |
| Ldiv48 | Ldiv58 | 0.248 | |
| Ldiv55 | Ldiv58 | 0.428 | |
3.5 Populations’ reproductive success
After eight weeks in the growth chamber, the higher seeds’ germinability was observed in population Camboazinho (40.67%), followed by Cacequi (32.67%), Canas (19.33%) and Inhatinhum with 18.00% (Table 3). However, population Cacequi revealed a much higher speed of germination (14.18) in comparison to the other ones (Table 3), presenting the faster seeds’ germination and emergence. In population Camboazinho, mean speed of germination was 9.20, in Inhatinhum it was 7.98 and in Canas it equaled 5.04.
| Table 3. Mean seeds’ germinability (%), speed of germination (mean ± SD), proportion of anomalous germinated seeds (%), inbreeding coefficient (f), and observed heterozygosity (HO) of L. divaricata in four populations within the Brazilian Pampa biome. Values of the germination traits are the mean for six mother-trees and 25 seeds/tree. The correlation (r) between each germination trait and the inbreeding coefficient and observed heterozygosity was estimated using the Pearson’s product-moment correlation coefficient. | |||||
| Population | Mean seeds’ germinability | Speed of germination | Anomalous germinated seeds | f a | HOa |
| Camboazinho | 40.67 | 8.81 ± 4.21 | 3.33 | 0.213 | 0.52 |
| Canas | 19.33 | 4.19 ± 4.75 | 6.67 | 0.221 | 0.57 |
| Inhatinhum | 18.00 | 6.96 ± 8.50 | 7.33 | 0.236 | 0.42 |
| Cacequi | 32.67 | 13.03 ± 3.67 | 4.67 | –0.017 | 0.54 |
| r (f) | –0.36 | –0.86 | 0.36 | ||
| r (HO) | 0.31 | 0.03 | –0.39 | ||
| a Data from Nagel et al. (2015) | |||||
The proportion of anomalous germinated plantlets ranged from 3.33% in Camboazinho to 7.33% in Inhatinhum (Table 3). The observed anomalies were the absence of one or both cotyledons, absence of the root systems and distorted growth of the plantlet.
A low correlation between seed’s germinability and the inbreeding coefficient (f), and observed heterozygosity (HO) was observed, being negative for f, but positive for HO (Table 3). The speed of germination revealed a high negative correlation (r = –0.86) with f and a very low positive correlation (r = 0.03) with the HO (Table 3). The proportion of anomalous germinated plantlets revealed low positive correlation with f, but negative with HO (Table 3).
4 Discussion
Molecular signatures of the occurrence of effective size reduction in the southern populations of L. divaricata were captured for all studied populations, with different results among the heterozygosity-excess, shifts in allele frequencies and the M-ratio tests. Such result is related to the difference of the timespan since the reduction of population effective size occurred. Shifts in allele size distribution and heterozygosity excess are transitory phenomena and may be erased in approximately 0.2–4Ne generations (Luikart and Cornuet 1998), while low M-ratios may persist for hundreds of generations (Garza and Williamson 2001). Considering an effective population size Ne = 100 individuals during the bottleneck and a generation interval of 20 years for L. divaricata, the tests employed in this study cover a timeframe from 400 years (0.2Ne generations) to 10 000 years (500 generations), enabling the capture of effective size reduction signatures within this timeframe.
Despite the lack of records about the historical census size of the studied populations, the palynological record of an adjacent site (Behling et al. 2005) suggests a colder and drier climate to this region up to about 5000 years before the present, making the demographic growth of forest species very hard. Large ice sheets or permafrost were not commonly observed in South America during the Quaternary ice ages (Hewitt 2000). However, the colder and drier climate (Behling 2002; Behling et al. 2005) likely led many plant species to experience significant reduction in effective population size, as suggested for Araucaria angustifolia populations from southern Brazil (Stefenon et al. 2008), and for a small isolated population of Pinus taeda in North America (Al-Rabab’ah and Williams 2004). In addition, ancient anthropic interference may not be discarded, since the earlier human inhabitants of this region are dated from this period (Behling et al. 2005; Schmitz 2006). Estimations of heterozygosity excess found in populations of L. divaricata from the Atlantic Forest were considered signatures of genetic bottlenecks originated from habitat loss and overharvesting (Conson et al. 2013), leading populations to isolation and fragmentation. Thus, the current genetic status of the species may be effected not only from climatic events from the Holocene, but may also have been influenced by ancient and recent human action. The action of indigenous people modifying the environment (e.g. cleaning the forest understory in the plantation of crops and dispersing seeds) was considered an important factor in shaping the current occurrence area and genetic structure of A. angustifolia in Southern Brazil (Stefenon et al. 2008).
The impact of the continuous isolation of small-sized forest formations may lead this species to loss of genetic diversity and adaptability potential within few generations due to inbreeding and genetic drift (Stefenon and Costa 2012). In small populations, the effects of genetic drift result in the consistent loss of rare allelic combinations, increasing linkage disequilibrium (LD) levels (Wang et al. 1998; Flint-Garcia et al. 2003). In our analysis, no pair of loci revealed significant LD in all populations, suggesting that the observed LD is not result of physical linkage, but caused by recent reduction in effective population size and accompanying genetic drift. During a bottleneck, only few allelic combinations are transmitted to future generations, causing substantial LD (Flint-Garcia et al. 2003). Accordingly, such loss of alleles was detected in the M-ratio analysis and in the heterozygosity-excess test for populations Camboazinho e Canas, which presented 50% and 70% of loci pairs with LD, respectively.
The founder effect of a bottleneck is expected to generate populations with low genetic diversity. The mean levels of genetic diversity overall loci estimated through number of alleles (![]() < 4.8 in four out of five populations) and expected heterozygosity under Hardy-Weinberg proportions (
< 4.8 in four out of five populations) and expected heterozygosity under Hardy-Weinberg proportions (![]() > 0.53) corroborates the expectation of bottlenecked populations that loss alleles with higher intensity than heterozygosity (Nei et al. 1975). In bottlenecked populations low frequency alleles are eliminated, reducing allelic richness, while the reduction in heterozygosity depends on the bottleneck size and on the rate of population growth after the bottleneck (Nei et al. 1975). Rapid population expansion may reduce the effects of the bottleneck, as suggested for bottlenecked populations of Pinus maximartinezii (Ledig et al. 1999), Pinus resinosa (Echt et al. 1998) and A. angustifolia (Stefenon et al. 2008). Similarly, L. divaricata populations may have experienced a relatively fast recovery of census size, accompanied by an increase of the expected heterozygosity (HE), without recuperation of the allelic richness.
> 0.53) corroborates the expectation of bottlenecked populations that loss alleles with higher intensity than heterozygosity (Nei et al. 1975). In bottlenecked populations low frequency alleles are eliminated, reducing allelic richness, while the reduction in heterozygosity depends on the bottleneck size and on the rate of population growth after the bottleneck (Nei et al. 1975). Rapid population expansion may reduce the effects of the bottleneck, as suggested for bottlenecked populations of Pinus maximartinezii (Ledig et al. 1999), Pinus resinosa (Echt et al. 1998) and A. angustifolia (Stefenon et al. 2008). Similarly, L. divaricata populations may have experienced a relatively fast recovery of census size, accompanied by an increase of the expected heterozygosity (HE), without recuperation of the allelic richness.
Such moderated levels of expected heterozygosity observed in populations of L. divaricata (Conson et al. 2013; Nagel et al. 2015) might also be effect of a high pre-bottleneck diversity. This moderate genetic diversity allowed the species extending over a large range of environments in South America across the Pampa, Atlantic Forest and Cerrado biomes and may be crucial for species survival under the current environmental changes.
Despite this moderate level of expected heterozygosity, the reproductive success of these populations may be under threat. The germinability (a measurement of germination capacity), the speed of germination (a measurement of the seedlings vigor) and the proportion of anomalous germinated plantlets seem to be at least partially affected by the levels of inbreeding. The observed correlation suggests that higher levels of inbreeding led to a decrease in populations’ fitness, decreasing germination capacity and seedling vigor, and increasing the levels of anomalous germinated plantlets. This occurs mostly due to the loss of rare alleles, fitting a model of founder event as proposed by Nagel et al. (2015). As reported for Gentiana pneumonanthe (Oostermeijer et al 1994) and Swainsona recta (Buza et al. 2000), the main concern regarding the increased level of gametic correlation is the reduction of individual fitness due to inbreeding depression and associated negative effects on the viability of small remnant populations. In addition, the potential for maternal plants in small populations to invest in offspring may also be reduced by inbreeding depression (Oostermeijer et al 1994).
Germination timing influences survival and may have important fitness consequences under environmental uncertainty (Simons and Johnston 2000), acting early in life history. Such early life history traits have been assumed to be under control of one or only a few loci (Husband and Schemske, 1996). Therefore, inbreeding depression in this case may occur due to a few recessive lethals. Usually, different populations tend to fix different random subsets of deleterious alleles that mask each other when populations are crossed (Keller and Waller 2002). Promoting connection among populations towards increasing populations’ heterozygosity and decreasing inbreeding as a strategy for populations conservation is supported by the results of Newman and Tallmon (2001), who demonstrated that one single migrant per generation can play an important role in preventing the harmful effects of inbreeding in small population and is sufficient to maintain genetic variation and fitness over short term. Accordingly, higher levels of observed heterozygosity in L. divaricata are correlated with populations’ fitness through higher germination capacity and lower proportion of anomalous germinated plantlets. Thus, considering the low allelic richness and observed heterozygosity, and the decrease of populations’ fitness, the promotion of connection among populations is an important strategy towards conservation of L. divaricata populations. The offspring from crosses among populations would significantly increase observed heterozygosity, decrease inbreeding and, consequently, increase populations’ fitness.
Acknowledgements
The Authors thank CNPq/Brazil (Processes 471812/2011-0 and 474758/2012-5) and UNIPAMPA (PROPESQ and PROPG) by the financial support. JCN received post-graduation scholarship by CAPES/Brazil.
References
Al-Rabab’ah M., Williams C.G. (2004). An ancient bottleneck in the Lost Pines of central Texas. Molecular Ecology 13(5): 1075–1084. http://dx.doi.org/10.1111/j.1365-294X.2004.02142.x.
Backes P., Irgang B. (2002). Trees from South: guide for identification and ecological interest. The main South Brazilian native species. Instituto Souza Cruz, São Paulo. 331 p. [In Portuguese].
Beaumont M.A. (1999). Detecting population expansion and decline using microsatellites. Genetics 153: 2013–2029.
Behling H. (2002). South and southeast Brazilian grasslands during Late Quaternary times: a synthesis. Palaeogeography, Palaeoclimatology, Palaeoecology 177(1–2): 19–27. http://dx.doi.org/10.1016/S0031-0182(01)00349-2.
Behling H., Pillar V.P., Bauermann S.G. (2005). Late Quaternary grassland (Campos), gallery forest, fire and climate dynamics, studied by pollen, charcoal and multivariate analysis of the São Francisco de Assis core in western Rio Grande do Sul (southern Brazil). Review of Palaeobotany and Palynology 133(3–4): 235–248. http://dx.doi.org/10.1016/j.revpalbo.2004.10.004.
Buza L., Young A., Thrall P. (2000). Genetic erosion, inbreeding and reduced fitness in fragmented populations of the endangered tetraploid pea Swainsona recta. Biological Conservation 93(2): 177–186. http://dx.doi.org/10.1016/S0006-3207(99)00150-0.
Carvalho P.E.R. (2003). Brazilain tree species. vol. 1. Embrapa Informação Tecnológica/Embrapa Florestas, Brasília, Colombo, Brazil. 1039 p. [In Portuguese].
Ceconi D.E., Poletto I., Brun E.J., Lovato T. (2006). Growth of seedlings of Açoita-cavalo (Luehea divaricata Mart.) under influence of phosphate fertilization. Cerne 12: 292–299. [In Portuguese].
Conson A.R.O, Ruas E.A., Vieira B.G., Rodrigues L.A., Costa B.F., Bianchini E., Prioli A.J., Ruas C.F., Ruas P.M. (2013). Genetic structure of the Atlantic Rainforest tree species Luehea divaricata (Malvaceae). Genetica 141(4): 205–215. http://dx.doi.org/10.1007/s10709-013-9719-4.
Cornuet J.M., Luikart G. (1996). Description and power of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144: 2001–2014.
Di Rienzo A., Peterson A.C., Garza J.C., Valdes A.M., Slatkin M., Freimer N.B. (1994). Mutational processes of simple-sequence repeat loci in human populations. Proceedings of the National Academy of Sciences USA 91(8): 3166–3170. http://dx.doi.org/10.1073/pnas.91.8.3166.
Echt C.S., Deverno L.L., Anziedei M., Vendramin G.G. (1998). Chloroplast microsatellites reveal population genetic diversity in red pine, Pinus resinosa Ait. Molecular Ecology 7(3): 307–316. http://dx.doi.org/10.1046/j.1365-294X.1998.00350.x.
Finkeldey R., Hattemer H.H. (2007). Tropical forest genetics. Springer Verlag, Berlin, Heidelberg. 316 p.
Flint-Garcia S.A., Thornsberry J.M., Buckler IV E.S. (2003). Structure of linkage disequilibrium in plants. Annual Reviews of Plant Biology 54: 357–374. http://dx.doi.org/10.1146/annurev.arplant.54.031902.134907.
Garza J.C., Williamson E.G. (2001). Detection of reduction in population size using data from microsatellite loci. Molecular Ecology 10: 305–318.
Hewitt G. (2000). The genetic legacy of the Quaternary ice ages. Nature 405: 907–913. http://dx.doi.org/10.1038/35016000.
Husband B., Schemske D. (1996). Evolution of the magnitude and timing of inbreeding depression in plants. Evolution 50(1): 54–70. http://dx.doi.org/10.2307/2410780.
Keller L.F., Waller D.M. (2002). Inbreeding effects in wild populations. Trends in Ecology and Evolution 17(5): 230–241. http://dx.doi.org/10.1016/S0169-5347(02)02489-8.
Kingman J.F.C. (1982). On the genealogy of large populations. Journal of Applied Probability 19A: 27–43.
Ledig F.T., Conkle M.-T., Velázquez B.B., Piedra T.E., Hodgskiss P.D., Johnson D.R., Dvorak W.S. (1999). Evidence for an extreme bottleneck in a rare Mexican pinyon: genetic diversity, disequilibrium, and the mating system in Pinus maximartinezzi. Evolution 53(1): 91–99. http://dx.doi.org/10.2307/2640922.
Luikart G., Cornuet J.-M. (1998). Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conservation Biology 12(1): 228–237. http://dx.doi.org/10.1111/j.1523-1739.1998.96388.x.
Luikart G., Sherwin W.B., Steele B.M., Allendorf F.W. (1998a). Usefulness of molecular markers for detecting population bottlenecks via monitoring genetic change. Molecular Ecology 7(8): 963–974. http://dx.doi.org/10.1046/j.1365-294x.1998.00414.x.
Luikart G., Allendorf F.W., Cornuet J.-M., Sherwin W.B. (1998b). Distortion of allele frequency distributions provides a test for recent population bottlenecks. Journal of Heredity 89(3): 238–247. http://dx.doi.org/10.1093/jhered/89.3.238.
Luijten S.H., Dierick A., Gerard J., Oostermeijer B., Raijmann L.E.L., Den Nijs H.C.M. (2000). Population size, genetic variation, and reproductive success in a rapidly declining, self-incompatible perennial (Arnica montana) in The Netherlands. Conservation Biology 14(6): 1776–1787. http://dx.doi.org/10.1111/j.1523-1739.2000.99345.x.
Maguire J.D. (1962). Speed of germination – aid in selection and evaluation for seedling emergence and vigor. Crop Science 2: 176–177.
Nagel J.C., Ceconi D.E., Poletto I., Stefenon V.M. (2015). Historical gene flow within and among populations of Luehea divaricata in the Brazilian Pampa. Genetica 143(3): 317–1329. http://dx.doi.org/10.1007/s10709-015-9830-9.
Nei M., Maruyama T., Chakraborty R. (1975). The bottleneck effect and genetic variability in populations. Evolution 29(1): 1–10. http://dx.doi.org/10.2307/2407137.
Newman D., Tallmon D.A. (2001) Experimental evidence for beneficial fitness effects of gene flow in recently isolated populations. Conservation Biology 15(4): 1054–1063. http://dx.doi.org/10.1046/j.1523-1739.2001.0150041054.x.
Ohta T., Kimura M. (1973). A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a genetic population. Genetics Research 22: 201–204. http://dx.doi.org/10.1017/S0016672308009531.
Oostermeijer J.G.B., van Eijck M.W., den Nijs J.C.M. (1994). Offspring fitness in relation to population size and genetic variation in the rare perennial plant species Gentiana pneumonanthe (Gentianaceae). Oecologia 97(3): 289–296. http://dx.doi.org/10.1007/BF00317317.
Paoli A.A.S. (1995). Morphology and development of seeds and plantlets of Luehea divaricata Mart. et Zucc. (Tiliaceae). Revista Brasileira de Sementes 17: 120–128. [In Portuguese].
Peery M.Z., Kirby R., Reid B.N., Stoelting R., Doucet-Bëer E., Robinson S., Vásquez-Carrillo C., Pauli J.N., Palsbøll P.J. (2012). Reliability of genetic bottleneck tests for detecting recent population declines. Molecular Ecology 21(14): 3403–3418. http://dx.doi.org/10.1111/j.1365-294X.2012.05635.x.
Piry S., Luikart G., Cornuet J.M. (1999). Bottleneck: a computer program for detecting recent reduction in the effective population size using allele frequency data. Journal of Heredity 90: 502–503. http://dx.doi.org/10.1093/jhered/90.4.502.
Ranal M.A., Santana D.G. (2006). How and why to measure the germination process? Revista Brasileira de Botânica 29: 1–11. http://dx.doi.org/10.1590/S0100-84042006000100002.
Raymond M., Rousset F. (1995). GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity 86: 248–249.
Reitz R., Klein R.M., Reis A. (1988). Project woods of the Rio Grande do Sul. Herbário Barbosa Rodrigues, Secretaria da Agricultura e Abastecimento. [In Portuguese].
Roesch L.F.W., Vieira F.C.B., Pereira V.A., Schünemann A.L., Teixeira I.F., Senna A.J.T., Stefenon V.M. (2009). The Brazilian Pampa: a fragile biome. Diversity 1(2): 182–198. http://dx.doi.org/10.3390/d1020182.
Ruas E.A., Conson A.R.O., Costa B.F., Damasceno J.O., Rodrigues L.A., Reck M., Vieira A.O.S., Ruas P.M., Ruas C.F. (2009). Isolation and characterization of tem microsatellite loci for the tree species Luehea divaricata Mart. (Malvaceae) and intergeneric transferability. Conservation Genetics Resources 1: 245–248. http://dx.doi.org/10.1007/s12686-009-9060-5.
Schmitz P.I. (2006). Archeology of the Rio Grande do Sul, Brazil. Instituto Anchietano de Pesquisas – UNISINOS, São Leopoldo. 164 p. [In Portuguese].
Simons A.M., Johnston M.O. (2000). Variation in seed traits of Lobelia inflata (Campanulaceae): sources and fitness consequences. American Journal of Botany 87(1): 124–132. http://dx.doi.org/10.2307/2656690.
Stefenon V.M., Behling H., Gailing O., Finkeldey R. (2008). Evidences of delayed size recovery in Araucaria angustifolia populations after post-glacial colonization of highlands in Southeastern Brazil. Anais da Academia Brasileira de Ciências 80: 433–443. http://dx.doi.org/10.1590/S0001-37652008000300005.
Stefenon V.M., Costa L.S. (2012). A simulation study on the behavior of allelic richness and inbreeding coefficient over generations in fragmented populations of tree species. Annals of Forest Research 55: 3–10.
Stefenon V.M., Nodari R.O. (2003). Molecular markers in the genetic breeding of Araucária. Biotecnologia Ciência e Desenvolvimento 31: 95–99. [In Portuguese].
Tanaka J.C.A., Silva C.C., Filho B.P.D., Nakamura C.V., Carvalho J.E., Foglio M.A. (2005). Chemical constituents of Luehea divaricata Mart. (Tiliaceae). Química Nova 28: 834–837. [In Portuguese].
Teixeira M.P., Cruz L., Franco J.L., Vieira R.B., Stefenon V.M. (2016). Ethnobotany and antioxidant evaluation of commercialized medicinal plants from the Brazilian Pampa. Acta Botanica Brasilica 30(1): 1–12. http://dx.doi.org/10.1590/0102-33062015abb0150.
Vigoroux Y., Jaqueth J.S., Matsuoka Y., Smith O.S., Beavis W.D., Stephen J., Smith C., Doebley J. (2002). Rate and pattern of mutation at microsatellite loci in maize. Molecular Biology and Evolution 19(8): 1251–1260. http://dx.doi.org/10.1093/oxfordjournals.molbev.a004186.
Wang J., Caballero A., Hill W.G. (1998). The effect of linkage disequilibrium and deviation from Hardy-Weinberg proportion on the changes in genetic variance with bottleneck. Heredity 81(2): 174–186. http://dx.doi.org/10.1046/j.1365-2540.1998.00390.x.
Wessa P. (2016). Free statistics software. Office for research development and education, version 1.1.23-r7. http://www.wessa.net/.
Total of 48 references.

